bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4723_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
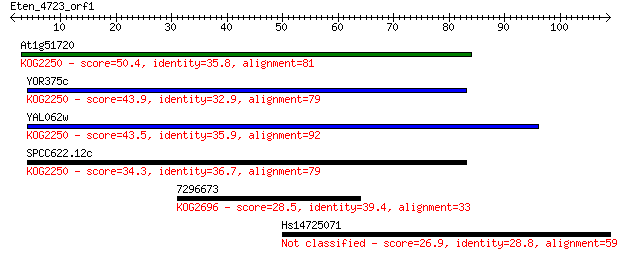
At1g51720 50.4 8e-07
YOR375c 43.9 8e-05
YAL062w 43.5 9e-05
SPCC622.12c 34.3 0.060
7296673 28.5 2.8
Hs14725071 26.9 9.7
> At1g51720
Length=624
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 44/81 (54%), Gaps = 1/81 (1%)
Query 3 EKLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFADGDSIVFQNS 62
EKL+A GA +T+SDS+G + GF + +++ K+ +R S + F +
Sbjct 421 EKLIACGAHPVTVSDSKGYLVDDDGFDYMKLAFLRDIKSQQRSLRDYSKTYARAKYF-DE 479
Query 63 ETPWAVAADLAFPCAKENEVD 83
PW D+AFPCA +NEVD
Sbjct 480 LKPWNERCDVAFPCASQNEVD 500
> YOR375c
Length=454
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 41/85 (48%), Gaps = 6/85 (7%)
Query 4 KLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEA----KAADSCVRVRSFADGDSIVF 59
K++ +G V++LSDS+G I G + E + I A K+ + V S + + +
Sbjct 235 KVIELGGTVVSLSDSKGCIISETGITSEQVADISSAKVNFKSLEQIVNEYSTFSENKVQY 294
Query 60 QNSETPWAVA--ADLAFPCAKENEV 82
PW D+A PCA +NEV
Sbjct 295 IAGARPWTHVQKVDIALPCATQNEV 319
> YAL062w
Length=457
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 52/105 (49%), Gaps = 18/105 (17%)
Query 4 KLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFAD--GDSIVFQN 61
K++ +G V++LSDS+G I G + E I I AK +R +S + + F
Sbjct 236 KVIELGGIVVSLSDSKGCIISETGITSEQIHDIASAK-----IRFKSLEEIVDEYSTFSE 290
Query 62 SET-------PWAVAA--DLAFPCAKENEV--DDIAAAMSSRAEF 95
S+ PW + D+A PCA +NEV D+ A ++S +F
Sbjct 291 SKMKYVAGARPWTHVSNVDIALPCATQNEVSGDEAKALVASGVKF 335
> SPCC622.12c
Length=451
Score = 34.3 bits (77), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 19/90 (21%)
Query 4 KLLAMGAKVLTLSDSEGMIY--CSQGFSKEDIQRI---KEAKAADSCVRVRSFADGDSIV 58
K + GA V ++SDS+G++ ++G E+I I KE +A S AD S+
Sbjct 238 KCIQEGAIVKSISDSKGVLIAKTAEGLVPEEIHEIMALKEKRA--------SIADSASLC 289
Query 59 FQNSET----PWAVAA--DLAFPCAKENEV 82
++ PW D+A PCA +NEV
Sbjct 290 KKHHYIAGARPWTNVGEIDIALPCATQNEV 319
> 7296673
Length=405
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 31 EDIQRIKEAKAADSCVRVRSFADGDSIVFQNSE 63
ED QR++ A SC++++SFA G+ + N E
Sbjct 267 EDFQRLRNFVDARSCMKLKSFAPGEIVKGFNKE 299
> Hs14725071
Length=1833
Score = 26.9 bits (58), Expect = 9.7, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query 50 SFADGDSIVFQNSETPWAVAADLAFPCAKENEVDDIAAAMSSRAEFGTRRERERERERE 108
S ++G VF + +TP + + F +E DD ++SS +E + E++ + ERE
Sbjct 515 SNSEGSCSVFSSPKTPGGFSPGIPFQ-TEEGRRDD---SLSSTSEDSEKDEKDEDHERE 569
Lambda K H
0.316 0.128 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160781780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40