bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4691_orf2
Length=241
Score E
Sequences producing significant alignments: (Bits) Value
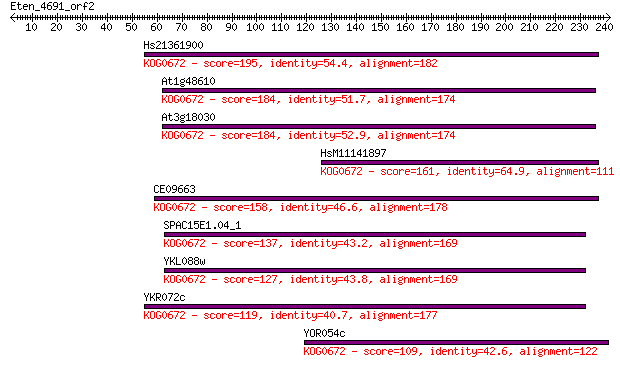
Hs21361900 195 8e-50
At1g48610 184 1e-46
At3g18030 184 2e-46
HsM11141897 161 1e-39
CE09663 158 1e-38
SPAC15E1.04_1 137 3e-32
YKL088w 127 2e-29
YKR072c 119 4e-27
YOR054c 109 4e-24
> Hs21361900
Length=204
Score = 195 bits (495), Expect = 8e-50, Method: Compositional matrix adjust.
Identities = 99/183 (54%), Positives = 127/183 (69%), Gaps = 7/183 (3%)
Query 55 PPAQGTVRVLLGVTGSVAAIKTAEVIQELHAAAAARGLTAEVDLVATEKGLHFL-PQPLR 113
P + VL+GVTGSVAA+K ++ +L GL EV +V TE+ HF PQ +
Sbjct 12 PLMERKFHVLVGVTGSVAALKLPLLVSKL---LDIPGL--EVSVVTTERAKHFYSPQDIP 66
Query 114 PLVRKDSQEWAAWKQRGDPVLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARA 173
+ D+ EW WK R DPVLHI+LRRWADL L+APL AN+L K++SG+CD+LL+CV RA
Sbjct 67 VTLYSDADEWEMWKSRSDPVLHIDLRRWADLLLVAPLDANTLGKVASGICDNLLTCVMRA 126
Query 174 WSYSSKPLVAAPAMNTEMWQHPITSQQLQQLQHFGAKIVPPVAKRLICGDTGVGAMAEPH 233
W SKPL+ PAMNT MW+HPIT+QQ+ QL+ FG +P VAK+L+CGD G+GAMAE
Sbjct 127 WD-RSKPLLFCPAMNTAMWEHPITAQQVDQLKAFGYVEIPCVAKKLVCGDEGLGAMAEVG 185
Query 234 TIA 236
TI
Sbjct 186 TIV 188
> At1g48610
Length=389
Score = 184 bits (468), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 90/177 (50%), Positives = 117/177 (66%), Gaps = 11/177 (6%)
Query 62 RVLLGVTGSVAAIKTAEVIQELHAAAAARGLTAEVDLVATEKGLHFLPQPLRP---LVRK 118
R+LL +GSVA+IK + + AEV VA++ L+F+ +P P +
Sbjct 201 RILLAASGSVASIKFSNLCHCFSE-------WAEVKAVASKSSLNFVDKPSLPQNVTLYT 253
Query 119 DSQEWAAWKQRGDPVLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARAWSYSS 178
D EW++W + GDPVLHIELRRWAD+ +IAPLSAN+LAKI+ GLCD+LL+C+ RAW Y S
Sbjct 254 DEDEWSSWNKIGDPVLHIELRRWADVMIIAPLSANTLAKIAGGLCDNLLTCIVRAWDY-S 312
Query 179 KPLVAAPAMNTEMWQHPITSQQLQQLQHFGAKIVPPVAKRLICGDTGVGAMAEPHTI 235
KPL APAMNT MW +P T + L L G ++PP+ K+L CGD G GAMAEP I
Sbjct 313 KPLFVAPAMNTLMWNNPFTERHLVLLDELGITLIPPIKKKLACGDYGNGAMAEPSLI 369
> At3g18030
Length=209
Score = 184 bits (466), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 92/177 (51%), Positives = 113/177 (63%), Gaps = 11/177 (6%)
Query 62 RVLLGVTGSVAAIKTAEVIQELHAAAAARGLTAEVDLVATEKGLHFLPQ---PLRPLVRK 118
RVLL +GSVAAIK + A EV V T+ LHFL + P +
Sbjct 21 RVLLAASGSVAAIKFGNLCHCFTEWA-------EVRAVVTKSSLHFLDKLSLPQEVTLYT 73
Query 119 DSQEWAAWKQRGDPVLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARAWSYSS 178
D EW++W + GDPVLHIELRRWAD+ +IAPLSAN+L KI+ GLCD+LL+C+ RAW Y +
Sbjct 74 DEDEWSSWNKIGDPVLHIELRRWADVLVIAPLSANTLGKIAGGLCDNLLTCIIRAWDY-T 132
Query 179 KPLVAAPAMNTEMWQHPITSQQLQQLQHFGAKIVPPVAKRLICGDTGVGAMAEPHTI 235
KPL APAMNT MW +P T + L L G ++PP+ KRL CGD G GAMAEP I
Sbjct 133 KPLFVAPAMNTLMWNNPFTERHLLSLDELGITLIPPIKKRLACGDYGNGAMAEPSLI 189
> HsM11141897
Length=127
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 72/111 (64%), Positives = 89/111 (80%), Gaps = 1/111 (0%)
Query 126 WKQRGDPVLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARAWSYSSKPLVAAP 185
WK R DPVLHI+LRRWADL L+APL AN+L K++SG+CD+LL+CV RAW SKPL+ P
Sbjct 2 WKSRSDPVLHIDLRRWADLLLVAPLDANTLGKVASGICDNLLTCVMRAWD-RSKPLLFCP 60
Query 186 AMNTEMWQHPITSQQLQQLQHFGAKIVPPVAKRLICGDTGVGAMAEPHTIA 236
AMNT MW+HPIT+QQ+ QL+ FG +P VAK+L+CGD G+GAMAE TI
Sbjct 61 AMNTAMWEHPITAQQVDQLKAFGYVEIPCVAKKLVCGDEGLGAMAEVGTIV 111
> CE09663
Length=237
Score = 158 bits (399), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 83/182 (45%), Positives = 112/182 (61%), Gaps = 7/182 (3%)
Query 59 GTVRVLLGVTGSVAAIKTAEVIQELHAAAAARGLTAEVDLVATEKGL---HFLPQPLRPL 115
G +LL +TGS+A +K E+I EL+ + +V V TE + H +
Sbjct 41 GKHNLLLILTGSIAVMKAPELISELYEKIGRDRILIKV--VTTENAMKLCHIQKLEFDEI 98
Query 116 VRKDSQEWAAWKQRGDPVLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARAWS 175
V +D EW+ W++RGD VLHIELR+WAD LIAPL AN++AKI++GLCD+L++ + RAW
Sbjct 99 VYEDRDEWSMWRERGDKVLHIELRKWADSALIAPLDANTMAKIANGLCDNLVTSIIRAWD 158
Query 176 YSSKPLVAAPAMNTEMWQHPITSQQLQQLQ-HFGAKIVPPVAKRLICGDTGVGAMAEPHT 234
SKP APAMNT MW++P+T Q L+ K + P+ K LICGD G GAMA T
Sbjct 159 L-SKPCYFAPAMNTHMWENPLTMQHRTVLKSQLKFKEICPIQKELICGDVGTGAMASIGT 217
Query 235 IA 236
I
Sbjct 218 IV 219
> SPAC15E1.04_1
Length=326
Score = 137 bits (344), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 73/175 (41%), Positives = 109/175 (62%), Gaps = 12/175 (6%)
Query 63 VLLGVTGSVAAIKTAEVIQELHAAAAARGLTAEVDLVATEKGLHFLPQP----LRPLVRK 118
+L+ TGSVAAIK +++ L +G+ +V +V T+ +F+ + L V
Sbjct 33 ILVAATGSVAAIKLTLIVKSL---LTYKGV--DVQVVLTDPARNFVEKEDLTALGVNVYN 87
Query 119 DSQEWAAWKQRGDPVLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARAWSYSS 178
++ +W W P+ HIELRRWA L LIAPLSAN++AK+++GLCD+LL+ + RAW+
Sbjct 88 NADDWKNWDGLECPITHIELRRWAHLLLIAPLSANTMAKMANGLCDNLLTSLIRAWA-PL 146
Query 179 KPLVAAPAMNTEMWQHPITSQQLQQLQHF--GAKIVPPVAKRLICGDTGVGAMAE 231
KP++ APAMNT MW +PIT + L + ++ + P+ K L CGD G+G MAE
Sbjct 147 KPILLAPAMNTLMWTNPITQEHLSAISRIYKNSEFIMPIEKVLACGDIGMGGMAE 201
> YKL088w
Length=571
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 74/181 (40%), Positives = 108/181 (59%), Gaps = 15/181 (8%)
Query 63 VLLGVTGSVAAIKTAEVIQELHAAAAARGLTAEVDLVATEKGLHFLPQPLRPLVRKDSQE 122
+L+G TGSVA IK +I +L ++ + L+ T+ HFL K +E
Sbjct 311 ILIGATGSVATIKVPLIIDKLFKIYGPEKIS--IQLIVTKPAEHFLKGLKMSTHVKIWRE 368
Query 123 WAAWK----QRGDP------VLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVAR 172
AW + D +LH ELR+WAD+FLIAPLSAN+LAK+++G+C++LL+ V R
Sbjct 369 EDAWVFDAVNKNDTSLSLNLILHHELRKWADIFLIAPLSANTLAKLANGICNNLLTSVMR 428
Query 173 AWSYSSKPLVAAPAMNTEMWQHPITSQQLQQL-QHF-GAKIVPPVAKRLICGDTGVGAMA 230
WS + P++ APAMNT M+ +P+T + L L Q + +++ PV K LICGD G+G M
Sbjct 429 DWSPLT-PVLIAPAMNTFMYINPMTKKHLTSLVQDYPFIQVLKPVEKVLICGDIGMGGMR 487
Query 231 E 231
E
Sbjct 488 E 488
> YKR072c
Length=562
Score = 119 bits (299), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 72/222 (32%), Positives = 108/222 (48%), Gaps = 48/222 (21%)
Query 55 PPAQGTVRVLLGVTGSVAAIKTAEVIQELHAAAAARGLTAEVDLVATEKGLHFLPQPLRP 114
P G + VL G TGS++ K +I++L ++ +V + T+ F Q
Sbjct 258 PQDDGKLHVLFGATGSLSVFKIKPMIKKLEEIYGRDRISIQV--ILTQSATQFFEQRYTK 315
Query 115 LVRK------------------------------------------DSQEWAAWKQRGDP 132
+ K D EW AWKQR DP
Sbjct 316 KIIKSSEKLNKMSQYESTPATPVTPTPGQCNMAQVVELPPHIQLWTDQDEWDAWKQRTDP 375
Query 133 VLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARAWSYSSKPLVAAPAMNTEMW 192
VLHIELRRWAD+ ++APL+AN+L+KI+ GLCD+LL+ V RAW+ S P++ AP+M + +
Sbjct 376 VLHIELRRWADILVVAPLTANTLSKIALGLCDNLLTSVIRAWN-PSYPILLAPSMVSSTF 434
Query 193 QHPITSQQLQQLQHFGA--KIVPPVAKRL-ICGDTGVGAMAE 231
+T +QLQ ++ + + P K + I GD G+G M +
Sbjct 435 NSMMTKKQLQTIKEEMSWVTVFKPSEKVMDINGDIGLGGMMD 476
> YOR054c
Length=674
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 52/125 (41%), Positives = 78/125 (62%), Gaps = 4/125 (3%)
Query 119 DSQEWAAWKQRGDPVLHIELRRWADLFLIAPLSANSLAKISSGLCDDLLSCVARAWSYSS 178
D EW W+QR DPVLHIELRRWAD+ ++APL+AN+LAKI+ GLCD+LL+ V RAW+ +
Sbjct 443 DQDEWDVWRQRTDPVLHIELRRWADILVVAPLTANTLAKIALGLCDNLLTSVIRAWN-PT 501
Query 179 KPLVAAPAMNTEMWQHPITSQQLQQLQHFG--AKIVPPVAKRL-ICGDTGVGAMAEPHTI 235
P+ AP+M + + +T + + +Q + P K + I GD G+ M + + I
Sbjct 502 FPIFLAPSMGSGTFNSIMTKKHFRIIQEEMPWVTVFKPSEKVMGINGDIGLSGMMDANEI 561
Query 236 ACEAL 240
+ +
Sbjct 562 VGKIV 566
Lambda K H
0.312 0.123 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4847568496
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40