bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4658_orf4
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
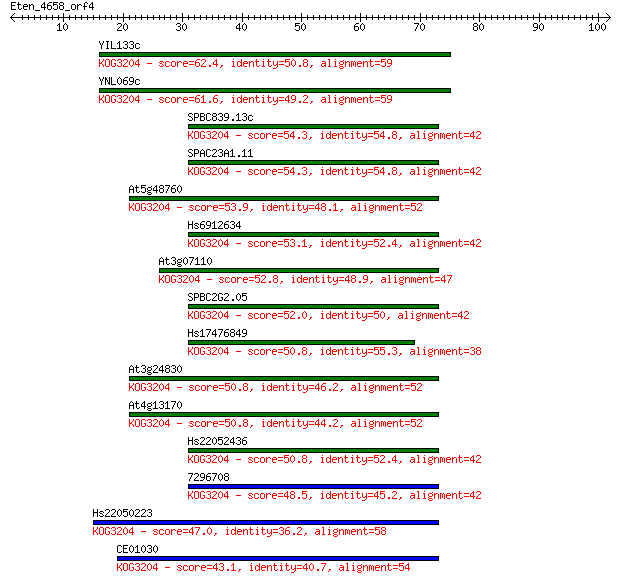
YIL133c 62.4 2e-10
YNL069c 61.6 4e-10
SPBC839.13c 54.3 5e-08
SPAC23A1.11 54.3 5e-08
At5g48760 53.9 8e-08
Hs6912634 53.1 1e-07
At3g07110 52.8 1e-07
SPBC2G2.05 52.0 3e-07
Hs17476849 50.8 6e-07
At3g24830 50.8 6e-07
At4g13170 50.8 6e-07
Hs22052436 50.8 7e-07
7296708 48.5 3e-06
Hs22050223 47.0 9e-06
CE01030 43.1 1e-04
> YIL133c
Length=199
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/61 (49%), Positives = 43/61 (70%), Gaps = 2/61 (3%)
Query 16 RQQQQQQMARDAAGV--LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSEL 73
R + AR A + L+V+EG+P Y++KK+VVVPQALRV+RL+PGR + LG+LS
Sbjct 85 RGMVSHKTARGKAALERLKVFEGIPPPYDKKKRVVVPQALRVLRLKPGRKYTTLGKLSTS 144
Query 74 I 74
+
Sbjct 145 V 145
> YNL069c
Length=198
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 43/61 (70%), Gaps = 2/61 (3%)
Query 16 RQQQQQQMARDAAGV--LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSEL 73
R + AR A + L+++EG+P Y++KK+VVVPQALRV+RL+PGR + LG+LS
Sbjct 84 RGMVSHKTARGKAALERLKIFEGIPPPYDKKKRVVVPQALRVLRLKPGRKYTTLGKLSTS 143
Query 74 I 74
+
Sbjct 144 V 144
> SPBC839.13c
Length=197
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 23/42 (54%), Positives = 33/42 (78%), Gaps = 0/42 (0%)
Query 31 LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
LQ EG+P ++++K+VVVP ALRV+RL+PGR +C +G LS
Sbjct 103 LQAVEGIPPPFDKQKRVVVPAALRVLRLKPGRKYCTVGRLSS 144
> SPAC23A1.11
Length=197
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 23/42 (54%), Positives = 33/42 (78%), Gaps = 0/42 (0%)
Query 31 LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
LQ EG+P ++++K+VVVP ALRV+RL+PGR +C +G LS
Sbjct 103 LQAVEGIPPPFDKQKRVVVPAALRVLRLKPGRKYCTVGRLSS 144
> At5g48760
Length=206
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Query 21 QQMARDAAGV--LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
+ R AA + L+VYEGVP Y++ K++V+P AL+V+RL+ G +C LG LS
Sbjct 95 HKTKRGAAALARLKVYEGVPTPYDKIKRMVIPDALKVLRLQAGHKYCLLGRLSS 148
> Hs6912634
Length=203
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 33/42 (78%), Gaps = 0/42 (0%)
Query 31 LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
L+V++G+P Y++KK++VVP AL+VVRL+P R F LG L+
Sbjct 102 LKVFDGIPPPYDKKKRMVVPAALKVVRLKPTRKFAYLGRLAH 143
> At3g07110
Length=206
Score = 52.8 bits (125), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 26 DAAGVLQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
+A L+V+EGVP Y++ K++VVP AL+V+RL+ G +C LG LS
Sbjct 102 NALARLKVFEGVPTPYDKIKRMVVPDALKVLRLQAGHKYCLLGRLSS 148
> SPBC2G2.05
Length=197
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 32/42 (76%), Gaps = 0/42 (0%)
Query 31 LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
LQ EG+P ++++K++VVP ALRV+RL+P R +C +G LS
Sbjct 103 LQALEGIPPPFDKQKRLVVPAALRVLRLKPSRKYCTIGRLSS 144
> Hs17476849
Length=359
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 21/38 (55%), Positives = 31/38 (81%), Gaps = 0/38 (0%)
Query 31 LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLG 68
L+V++G+P Y++KK++VVP AL+VVRL+P R F LG
Sbjct 268 LKVFDGIPPPYDKKKRMVVPAALKVVRLKPTRKFALLG 305
> At3g24830
Length=206
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Query 21 QQMARDAAGV--LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
+ R AA + L+V+EGVP Y++ K++V+P AL+V+RL+ G +C LG LS
Sbjct 95 HKTKRGAAALARLKVFEGVPPPYDKVKRMVIPDALKVLRLQAGHKYCLLGRLSS 148
> At4g13170
Length=206
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Query 21 QQMARDAAGV--LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
+ R AA + L+V+EG+P Y++ K++V+P AL+V+RL+ G +C LG LS
Sbjct 95 HKTKRGAAALARLKVFEGIPPPYDKIKRMVIPDALKVLRLQSGHKYCLLGRLSS 148
> Hs22052436
Length=203
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 32/42 (76%), Gaps = 0/42 (0%)
Query 31 LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
L+V +G+P Y++KK++VVP AL+VVRL+P R F LG L+
Sbjct 102 LKVSDGIPPPYDKKKRMVVPAALKVVRLKPTRKFAYLGRLAH 143
> 7296708
Length=205
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 33/42 (78%), Gaps = 0/42 (0%)
Query 31 LQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
L+V++G+P Y+++++VVVP A+RV+ LR R +C++G LS
Sbjct 104 LRVFDGIPSPYDKRRRVVVPIAMRVLTLRSDRKYCQVGRLSH 145
> Hs22050223
Length=154
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 36/58 (62%), Gaps = 0/58 (0%)
Query 15 RRQQQQQQMARDAAGVLQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
R Q Q + + L+V++G P Y++KK++++P AL+V+RL+ R F LG L+
Sbjct 37 RHAASQDQGGQASLDCLKVFDGTPPPYDKKKQMMIPAALKVMRLKLTRKFVYLGHLAH 94
> CE01030
Length=202
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 19 QQQQMARDAAGVLQVYEGVPVKYERKKKVVVPQALRVVRLRPGRDFCRLGELSE 72
+ +A L+ YEGVP KY++ K + P A R RL+P R FC +G LS
Sbjct 91 HKTNRGNEALKNLRAYEGVPAKYQKTKSLHAPSASR-FRLQPRRKFCVVGRLSH 143
Lambda K H
0.322 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40