bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
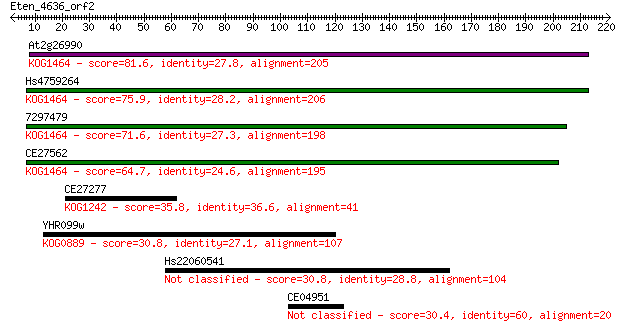
Query= Eten_4636_orf2
Length=220
Score E
Sequences producing significant alignments: (Bits) Value
At2g26990 81.6 1e-15
Hs4759264 75.9 6e-14
7297479 71.6 1e-12
CE27562 64.7 1e-10
CE27277 35.8 0.075
YHR099w 30.8 2.2
Hs22060541 30.8 2.3
CE04951 30.4 2.6
> At2g26990
Length=433
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 57/213 (26%), Positives = 97/213 (45%), Gaps = 32/213 (15%)
Query 8 QLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWKL 62
+L ++ +GE+ + +L +H++ G D Q LE A+E+ T +D K
Sbjct 144 KLCNIWFDIGEYRRMTKILKELHKSCQKEDGTDDQKKGSQLLEVYAIEIQIYTETKDNKK 203
Query 63 LQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQP 122
L+QL AL + +A PR +RECG G+ + +RQ
Sbjct 204 LKQLYHKALAIKSAIPHPRIMGIIRECG--------GKMHMAERQ--------------- 240
Query 123 WTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDVQ 182
W + AF+++ E G+ R L+ VL+ L S+V+PF +E K + DP++
Sbjct 241 WEEAATDFFEAFKNYDEAGNQR-RIQCLKYLVLANMLMESEVNPFDGQEAKPYKNDPEIL 299
Query 183 SAKALRAAFEANDVAGVEKHLQVLESV---DPF 212
+ L AA++ N++ E+ L+ DPF
Sbjct 300 AMTNLIAAYQRNEIIEFERILKSNRRTIMDDPF 332
> Hs4759264
Length=443
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 58/214 (27%), Positives = 98/214 (45%), Gaps = 32/214 (14%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +LY++ E+ K + +L +HQ+ G D Q LE A+E+ T ++ K
Sbjct 153 TKLGKLYLEREEYGKLQKILRQLHQSCQTDDGEDDLKKGTQLLEIYALEIQMYTAQKNNK 212
Query 62 LLQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQ 121
L+ L +L + +A P +RECG ++ H R E E
Sbjct 213 KLKALYEQSLHIKSAIPHPLIMGVIRECGGKM----HLR--------------EGE---- 250
Query 122 PWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDV 181
+ H AF+++ E G R L+ VL+ L S ++PF ++E K + DP++
Sbjct 251 -FEKAHTDFFEAFKNYDESGSPRRTTC-LKYLVLANMLMKSGINPFDSQEAKPYKNDPEI 308
Query 182 QSAKALRAAFEANDVAGVEKHLQVLESV---DPF 212
+ L +A++ ND+ EK L+ S DPF
Sbjct 309 LAMTNLVSAYQNNDITEFEKILKTNHSNIMDDPF 342
> 7297479
Length=444
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 54/203 (26%), Positives = 94/203 (46%), Gaps = 29/203 (14%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGVHQA-----GRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +LY ++TK + +L +HQ+ G D Q LE A+E+ T ++ K
Sbjct 154 TKLGKLYFDRSDFTKLQKILKQLHQSCQTDDGEDDLKKGTQLLEIYALEIQMYTVQKNNK 213
Query 62 LLQQLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQHQ 121
L+ L +L + +A P +RECG ++ H R + E E+ H
Sbjct 214 KLKALYEQSLHIKSAIPHPLIMGVIRECGGKM----HLR------------EGEFEKAH- 256
Query 122 PWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGDPDV 181
TD AF+++ E G R L+ VL+ L S ++PF ++E K + DP++
Sbjct 257 --TD----FFEAFKNYDESGSPRRTTC-LKYLVLANMLMKSGINPFDSQEAKPYKNDPEI 309
Query 182 QSAKALRAAFEANDVAGVEKHLQ 204
+ L +++ ND+ E L+
Sbjct 310 LAMTNLVNSYQNNDINEFETILR 332
> CE27562
Length=495
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 48/203 (23%), Positives = 91/203 (44%), Gaps = 32/203 (15%)
Query 7 SQLLRLYMQLGEWTKAEDLLPGV-----HQAGRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
++L +L+ L E+TK E ++ + ++ G + Q LE A+E+ T ++ K
Sbjct 150 TKLGKLFFDLHEFTKLEKIVKQLKVSCKNEQGEEDQRKGTQLLEIYALEIQMYTEQKNNK 209
Query 62 LLQ---QLLPHALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEE 118
L+ +L A+ +A P +RECG ++ + GR
Sbjct 210 ALKWVYELATQAIHTKSAIPHPLILGTIRECGGKMHLR-DGR------------------ 250
Query 119 QHQPWTDIHAHLMAAFRHHQEIGDARAAAAALRRAVLSAPLAGSDVDPFSTREGKALQGD 178
+ D H AF+++ E G R L+ VL+ L SD++PF ++E K + +
Sbjct 251 ----FLDAHTDFFEAFKNYDESGSPRRTTC-LKYLVLANMLIKSDINPFDSQEAKPFKNE 305
Query 179 PDVQSAKALRAAFEANDVAGVEK 201
P++ + + A++ ND+ E+
Sbjct 306 PEIVAMTQMVQAYQDNDIQAFEQ 328
> CE27277
Length=1788
Score = 35.8 bits (81), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 21 KAEDLLPGVHQAGRDPSLPPHQQLEAAAVEVLCCTYGRDWK 61
K DLL +H+ D P H++ A+E+LCCT G+ ++
Sbjct 537 KDTDLLGSIHKNMEDKKSPKHREGGLLALEILCCTIGKLFE 577
> YHR099w
Length=3744
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 29/110 (26%), Positives = 41/110 (37%), Gaps = 14/110 (12%)
Query 13 YMQLGEWTKAEDLLPGVHQAGRDPSLPPHQQLEAAAVE---VLCCTYGRDWKLLQQLLPH 69
Y Q+G W KA+ L R +L P+ Q E A E + C + W +L +L H
Sbjct 2694 YEQIGLWDKAQQLYEVAQVKARSGAL-PYSQSEYALWEDNWIQCAEKLQHWDVLTELAKH 2752
Query 70 ALRLSAASLDPRSAAAVRECGARLLIELHGRSDLQQRQQQQNDQQEPEEQ 119
+ ECG R+ R L+Q + D P Q
Sbjct 2753 E----------GFTDLLLECGWRVADWNSDRDALEQSVKSVMDVPTPRRQ 2792
> Hs22060541
Length=289
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 48/120 (40%), Gaps = 16/120 (13%)
Query 58 RDWKLLQQLLPHALRLSAASLD-PRSAAAVRECGARLLI-ELHGRSDLQQRQQQQNDQQE 115
RDW+LLQ LL A L A+ + + + E L ++ ++ Q QN +
Sbjct 107 RDWELLQSLLEQATSLRASRCEQVETVCGITEVQGHFLSGQVSATTECPQIAGLQNSSEF 166
Query 116 PEEQHQ-----PWTDIHAH-----LMAAFRHH----QEIGDARAAAAALRRAVLSAPLAG 161
+ + W D A R+H Q +GD ++ + L A LS P +G
Sbjct 167 GKRRKALTSVPKWEDTTASYPSTAYTPTLRYHPLVSQTLGDTSSSPSVLFEAPLSVPTSG 226
> CE04951
Length=2215
Score = 30.4 bits (67), Expect = 2.6, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 103 LQQRQQQQNDQQEPEEQHQP 122
LQ + Q N QQ+PE QHQP
Sbjct 1673 LQDHEHQYNSQQQPEYQHQP 1692
Lambda K H
0.319 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4147789496
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40