bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4562_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
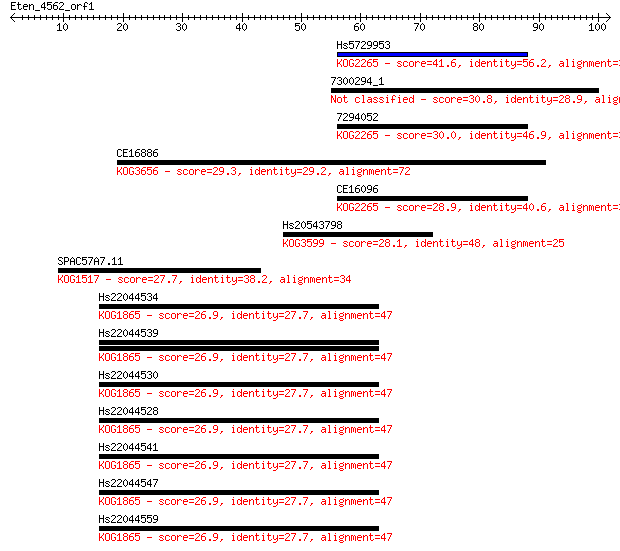
Hs5729953 41.6 4e-04
7300294_1 30.8 0.67
7294052 30.0 0.97
CE16886 29.3 1.8
CE16096 28.9 2.6
Hs20543798 28.1 4.5
SPAC57A7.11 27.7 4.8
Hs22044534 26.9 8.5
Hs22044539 26.9 8.6
Hs22044530 26.9 8.6
Hs22044528 26.9 8.6
Hs22044541 26.9 9.1
Hs22044547 26.9 9.2
Hs22044559 26.9 9.6
> Hs5729953
Length=331
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 18/33 (54%), Positives = 27/33 (81%), Gaps = 1/33 (3%)
Query 56 EGLLLALARGHDG-IGPLLDTFFSFLASRTDFF 87
+G+LLA+A+ H+G + L++TFFSFL +TDFF
Sbjct 11 DGMLLAMAQQHEGGVQELVNTFFSFLRRKTDFF 43
> 7300294_1
Length=403
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 22/45 (48%), Gaps = 0/45 (0%)
Query 55 HEGLLLALARGHDGIGPLLDTFFSFLASRTDFFHILEPGGPSMGF 99
++ +L+ + + I LD+ F FL TDF+H +GF
Sbjct 291 NDAMLMEILQDRKTITGFLDSIFGFLRRNTDFYHTKRDEADKIGF 335
> 7294052
Length=332
Score = 30.0 bits (66), Expect = 0.97, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Query 56 EGLLLALA-RGHDGIGPLLDTFFSFLASRTDFF 87
+ +LLA+A + H G+ L T SFL +TDFF
Sbjct 9 DNILLAVAEKHHGGVPEFLGTLASFLRRKTDFF 41
> CE16886
Length=395
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 3/75 (4%)
Query 19 LCVLLLHCVSDFHSPSHLVEVACIAAKFDFFFSKMDHEGLLLALARGH---DGIGPLLDT 75
L + ++ V +P++ V+ A+ FFF+ M HE + L H +G +L T
Sbjct 52 LVITVVLKVRGMKTPTNCYLVSLAASDTLFFFASMPHEMMYLLGPNDHYLFGSLGCVLLT 111
Query 76 FFSFLASRTDFFHIL 90
+ +LA T IL
Sbjct 112 YLPYLAMNTSSLSIL 126
> CE16096
Length=320
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Query 56 EGLLLALARGHDGIGP-LLDTFFSFLASRTDFF 87
+ +LL++A+ G P +LD F FL+ +TDF+
Sbjct 8 DSVLLSMAQQLSGGVPEMLDVLFEFLSRKTDFY 40
> Hs20543798
Length=945
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 47 DFFFSKMDHEGLLLALARGHDGIGP 71
D FF + H G L + GHDG+GP
Sbjct 92 DTFFLEAVHLGDLCKIVIGHDGLGP 116
> SPAC57A7.11
Length=1313
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 1/35 (2%)
Query 9 GVPSCEIACCLCVLLLHCVSDFHSPSHLV-EVACI 42
G P ++AC +L HC+S +SP L+ + AC+
Sbjct 609 GFPQGQLACLNPQVLSHCLSHLNSPDSLLRQWACL 643
> Hs22044534
Length=530
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 122 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 169
> Hs22044539
Length=1074
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 122 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 169
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 666 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 713
> Hs22044530
Length=565
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 157 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 204
> Hs22044528
Length=446
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 38 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 85
> Hs22044541
Length=525
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 117 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 164
> Hs22044547
Length=530
Score = 26.9 bits (58), Expect = 9.2, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 122 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 169
> Hs22044559
Length=530
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 16 ACCLCVLLLHCVSDFHSPSHLVEVA-CIAAKFDFFFSKMDHEGLLLAL 62
C LC + H H+P H+++ + +AA F + HE L+ +
Sbjct 122 GCMLCTMQAHITRALHNPGHVIQPSQALAAGFHRGKQEDAHEFLMFTV 169
Lambda K H
0.332 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40