bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4467_orf1
Length=56
Score E
Sequences producing significant alignments: (Bits) Value
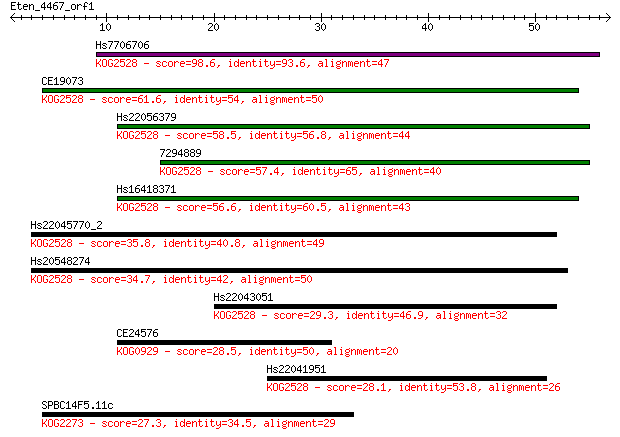
Hs7706706 98.6 3e-21
CE19073 61.6 4e-10
Hs22056379 58.5 3e-09
7294889 57.4 7e-09
Hs16418371 56.6 1e-08
Hs22045770_2 35.8 0.018
Hs20548274 34.7 0.045
Hs22043051 29.3 1.7
CE24576 28.5 2.8
Hs22041951 28.1 3.8
SPBC14F5.11c 27.3 6.2
> Hs7706706
Length=595
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 44/47 (93%), Positives = 46/47 (97%), Gaps = 0/47 (0%)
Query 9 EAWMTRMCRHPVISESEVFQQFLNFRDEKEWKTGKRKAEKDELGGVM 55
+AWMTRMCRHPVISESEVFQQFLNFRDEKEWKTGKRKAE+DEL GVM
Sbjct 332 QAWMTRMCRHPVISESEVFQQFLNFRDEKEWKTGKRKAERDELAGVM 378
> CE19073
Length=361
Score = 61.6 bits (148), Expect = 4e-10, Method: Composition-based stats.
Identities = 27/51 (52%), Positives = 39/51 (76%), Gaps = 1/51 (1%)
Query 4 RNSAPEAWMTRMCRHPVISESEVFQQFLNFRDEKEWKTGKRKAEKDE-LGG 53
R + W+ ++CRHPV+S+SEV+ F++ DEK+WK GKR+AEKDE +GG
Sbjct 93 RKHILQLWVNKICRHPVLSQSEVWLHFISCTDEKDWKNGKRRAEKDEYIGG 143
> Hs22056379
Length=574
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 25/44 (56%), Positives = 32/44 (72%), Gaps = 0/44 (0%)
Query 11 WMTRMCRHPVISESEVFQQFLNFRDEKEWKTGKRKAEKDELGGV 54
WM M HPV+S+ E FQ FL+ D+K+WK GKR+AEKDE+ G
Sbjct 313 WMDHMTSHPVLSQYEGFQHFLSCLDDKQWKMGKRRAEKDEMVGA 356
> 7294889
Length=531
Score = 57.4 bits (137), Expect = 7e-09, Method: Composition-based stats.
Identities = 26/40 (65%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 15 MCRHPVISESEVFQQFLNFRDEKEWKTGKRKAEKDELGGV 54
+CRHPVIS+ EV+ FL RDEK WK+GKRKAE+D GV
Sbjct 272 VCRHPVISKCEVWYHFLTCRDEKIWKSGKRKAERDPYMGV 311
> Hs16418371
Length=628
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/46 (56%), Positives = 32/46 (69%), Gaps = 3/46 (6%)
Query 11 WMTRMCRHPVISESEVFQQFL---NFRDEKEWKTGKRKAEKDELGG 53
WM M HPV+++ +VFQ FL + DEK WK GKRKAEKDE+ G
Sbjct 359 WMNHMASHPVLAQCDVFQHFLTCPSSTDEKAWKQGKRKAEKDEMVG 404
> Hs22045770_2
Length=150
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 3 CRNSAPEAWMTRMCRHPVISESEVFQQFL---NFRDEKEWKTGKRKAEKDEL 51
CR P +WM M V ++ VF F+ +E W+ GKRKA KDE+
Sbjct 5 CRK-GPISWMNHMASFWVQAQCHVFHHFVMCHTSTEETAWRQGKRKAAKDEV 55
> Hs20548274
Length=489
Score = 34.7 bits (78), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 26/53 (49%), Gaps = 4/53 (7%)
Query 3 CRNSAPEAWMTRMCRHPVISESEVFQQFL---NFRDEKEWKTGKRKAEKDELG 52
CR P WM + ++ +F FL E W+ GKRKAEKDELG
Sbjct 323 CRK-GPIWWMNHTASFWMQAQCHIFCHFLMRPTSTKETAWRQGKRKAEKDELG 374
> Hs22043051
Length=226
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 3/35 (8%)
Query 20 VISESEVFQQFL---NFRDEKEWKTGKRKAEKDEL 51
V ++ +F FL +E W+ GKRKAEKDE+
Sbjct 95 VQAQCHIFCHFLMCPTSTEETAWRQGKRKAEKDEM 129
> CE24576
Length=1628
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 10/20 (50%), Positives = 13/20 (65%), Gaps = 0/20 (0%)
Query 11 WMTRMCRHPVISESEVFQQF 30
WM+ C H ++S EVF QF
Sbjct 1290 WMSTTCNHAMLSVVEVFTQF 1309
> Hs22041951
Length=231
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 17/29 (58%), Gaps = 3/29 (10%)
Query 25 EVFQQFL---NFRDEKEWKTGKRKAEKDE 50
+F FL +E W+ GKRKAEKDE
Sbjct 93 HIFCHFLMCPTSTEETAWRQGKRKAEKDE 121
> SPBC14F5.11c
Length=586
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 4 RNSAPEAWMTRMCRHPVISESEVFQQFLN 32
R + ++ R+ +HP++ SEVF++FL+
Sbjct 152 RKRLLQLFLRRVAQHPILGLSEVFRKFLS 180
Lambda K H
0.317 0.133 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194096762
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40