bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
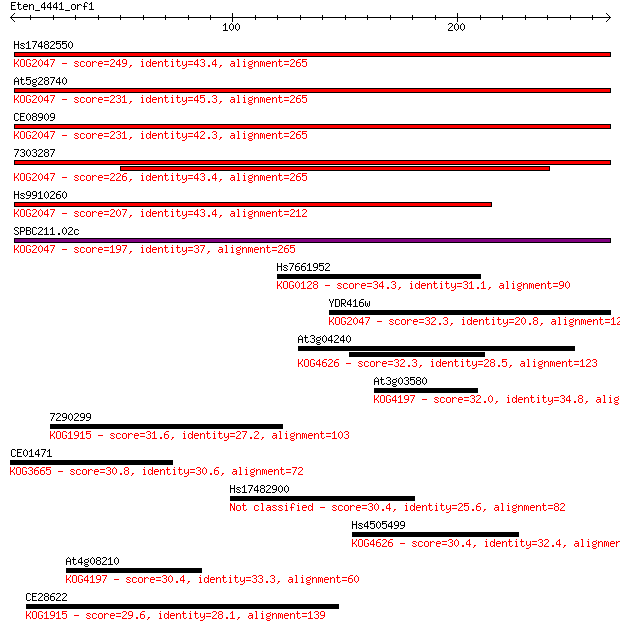
Query= Eten_4441_orf1
Length=267
Score E
Sequences producing significant alignments: (Bits) Value
Hs17482550 249 5e-66
At5g28740 231 1e-60
CE08909 231 2e-60
7303287 226 4e-59
Hs9910260 207 2e-53
SPBC211.02c 197 1e-50
Hs7661952 34.3 0.30
YDR416w 32.3 1.1
At3g04240 32.3 1.2
At3g03580 32.0 1.5
7290299 31.6 1.9
CE01471 30.8 3.5
Hs17482900 30.4 3.5
Hs4505499 30.4 3.7
At4g08210 30.4 4.0
CE28622 29.6 6.0
> Hs17482550
Length=855
Score = 249 bits (635), Expect = 5e-66, Method: Compositional matrix adjust.
Identities = 115/265 (43%), Positives = 172/265 (64%), Gaps = 2/265 (0%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R V + +MP +W +FL Q +T RRTFD ALRAL +TQH +WP + +++S
Sbjct 101 RAFVFMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFLRSHP 160
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PETAV YRR++ + PE +E+ YL S R DEAA LA V +D + +G++ ++L
Sbjct 161 LPETAVRGYRRFLKLSPESAEEYIEYLKSSDRLDEAAQRLATVVNDERFVSKAGKSNYQL 220
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W ELC L++ +P+ + S+ V+ I+R G+ RF+D + LWCSLA +Y+R G KARD+YE
Sbjct 221 WHELCDLISQNPDKVQSLNVDAIIRGGLTRFTDQLGKLWCSLADYYIRSGHFEKARDVYE 280
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLERLT 242
EA+ +V T+RD V+++YA FEE+++A ++ + E D++ +AR E+L
Sbjct 281 EAIRTVMTVRDFTQVFDSYAQFEESMIAAKMETASELGREEEDDV--DLELRLARFEQLI 338
Query 243 XRRPLLISSCKLRQNPHNVHEWLAR 267
RRPLL++S LRQNPH+VHEW R
Sbjct 339 SRRPLLLNSVLLRQNPHHVHEWHKR 363
> At5g28740
Length=917
Score = 231 bits (589), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 120/299 (40%), Positives = 170/299 (56%), Gaps = 34/299 (11%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R LV + +MP IW M L+ L QQL+TR RRTFD AL AL VTQH+ +W + +V G
Sbjct 93 RGLVTMHKMPRIWVMYLQTLTVQQLITRTRRTFDRALCALPVTQHDRIWEPYLVFVSQNG 152
Query 63 VP-ETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHE 121
+P ET++ +YRR++M +P +EF +L R+ E+A LA+V +D + G+T+H+
Sbjct 153 IPIETSLRVYRRYLMYDPSHIEEFIEFLVKSERWQESAERLASVLNDDKFYSIKGKTKHK 212
Query 122 LWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIY 181
LWLELC+L+ H I + V+ I+R GI +F+D V LW SLA +Y+R LL KARDIY
Sbjct 213 LWLELCELLVHHANVISGLNVDAIIRGGIRKFTDEVGMLWTSLADYYIRKNLLEKARDIY 272
Query 182 EEAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAA------------- 228
EE + V T+RD +V+++ Y+ FEE+ VA+ ++ + E
Sbjct 273 EEGMMKVVTVRDFSVIFDVYSRFEESTVAKKMEMMSSSDEEDENEENGVEDDEEDVRLNF 332
Query 229 --------------------KDIDFAIARLERLTXRRPLLISSCKLRQNPHNVHEWLAR 267
D+D +ARLE L RRP L +S LRQNPHNV +W R
Sbjct 333 NLSVKELQRKILNGFWLNDDNDVDLRLARLEELMNRRPALANSVLLRQNPHNVEQWHRR 391
> CE08909
Length=855
Score = 231 bits (588), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/266 (42%), Positives = 167/266 (62%), Gaps = 5/266 (1%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
RCL+ L +MP IW + + ++ L+T RR FD ALR+L VTQH +W + ++ S
Sbjct 108 RCLMRLHKMPRIWICYCEVMIKRGLITETRRVFDRALRSLPVTQHMRIWTLYIGFLTSHD 167
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PET + +YRR++ + P+ +++ YL + DEAA L + + + GRT H+L
Sbjct 168 LPETTIRVYRRYLKMNPKAREDYVEYLIERDQIDEAAKELTTLVNQDQNVSEKGRTAHQL 227
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W +LC L++ +P I S+ V+ I+R GI R++D V LWCSLA +Y+R +ARD+YE
Sbjct 228 WTQLCDLISKNPVKIFSLNVDAIIRQGIYRYTDQVGFLWCSLADYYIRSAEFERARDVYE 287
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQE-QQQASPEAAAAAAKDIDFAIARLERL 241
EA++ VST+RD A VY+AYAAFEE V+ ++QE +Q PE D+++ R + L
Sbjct 288 EAIAKVSTVRDFAQVYDAYAAFEEREVSIMMQEVEQSGDPEEEV----DLEWMFQRYQHL 343
Query 242 TXRRPLLISSCKLRQNPHNVHEWLAR 267
R+ L++S LRQNPHNV EWL R
Sbjct 344 MERKNELMNSVLLRQNPHNVGEWLNR 369
> 7303287
Length=883
Score = 226 bits (576), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 115/265 (43%), Positives = 167/265 (63%), Gaps = 1/265 (0%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R LV + +MP IW F+ Q +TR R FD ALRAL +TQH +WP + +V+
Sbjct 99 RALVFMHKMPRIWMDYGAFMTSQCKITRTRHVFDRALRALPITQHGRIWPLYLQFVRRFE 158
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PETA+ +YRR++ + PE T+E+ YL R DEAA LA + + + G++ H+L
Sbjct 159 MPETALRVYRRYLKLFPEDTEEYVDYLQEADRLDEAAQQLAHIVDNEHFVSKHGKSNHQL 218
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W ELC L++ +P +HS+ V+ I+R G+ R++D + LW SLA +YVR GL +ARDIYE
Sbjct 219 WNELCDLISKNPHKVHSLNVDAIIRGGLRRYTDQLGHLWNSLADYYVRSGLFDRARDIYE 278
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLERLT 242
EA+ +V+T+RD V++ YA FEE + + + EQ A+ A D++ ++R E L
Sbjct 279 EAIQTVTTVRDFTQVFDEYAQFEELSLNRRM-EQVAANEAATEEDDIDVELRLSRFEYLM 337
Query 243 XRRPLLISSCKLRQNPHNVHEWLAR 267
RR LL++S LRQNPHNVHEW R
Sbjct 338 ERRLLLLNSVLLRQNPHNVHEWHKR 362
Score = 30.0 bits (66), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 56/227 (24%), Positives = 89/227 (39%), Gaps = 37/227 (16%)
Query 50 LWPEMMDYVKSCGVPETAVCLYRRWIMIEP------ERTDEFALY--LSSIGRYDEAASH 101
LW + DY G+ + A +Y I + DE+A + LS R ++ A++
Sbjct 256 LWNSLADYYVRSGLFDRARDIYEEAIQTVTTVRDFTQVFDEYAQFEELSLNRRMEQVAAN 315
Query 102 LAAVASDSTITTASGRTRHELWLELCKLVAA------HPESIHS--MRV---EDILRSGI 150
AA D I +R E +E L+ +P ++H RV ED I
Sbjct 316 EAATEEDD-IDVELRLSRFEYLMERRLLLLNSVLLRQNPHNVHEWHKRVTLYEDKPAEII 374
Query 151 ARFSDAVAS------------LWCSLASHYVRMGLLAKARDIYEEAVS-SVSTIRDLAVV 197
+ +++AV + LW A Y G + AR ++E + DLA V
Sbjct 375 STYTEAVQTVQPKQAVGKLHTLWVEFAKFYEANGQVEDARVVFERGTEVEYVKVEDLAAV 434
Query 198 YEAYAAFE----EAVVAQLLQEQQQASPEAAAAAAKDIDFAIARLER 240
+ +A E + A L ++ A P+ A D + ARL R
Sbjct 435 WCEWAEMELRQQQFEAALKLMQRATAMPKRKIAYYDDTETVQARLHR 481
> Hs9910260
Length=855
Score = 207 bits (528), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 92/212 (43%), Positives = 141/212 (66%), Gaps = 0/212 (0%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R V + +MP +W +FL Q +T RRTFD ALRAL +TQH +WP + +++S
Sbjct 101 RAFVFMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFLRSHP 160
Query 63 VPETAVCLYRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHEL 122
+PETAV YRR++ + PE +E+ YL S R DEAA LA V +D + +G++ ++L
Sbjct 161 LPETAVRGYRRFLKLSPESAEEYIEYLKSSDRLDEAAQRLATVVNDERFVSKAGKSNYQL 220
Query 123 WLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYE 182
W ELC L++ +P+ + S+ V+ I+R G+ RF+D + LWCSLA +Y+R G KARD+YE
Sbjct 221 WHELCDLISQNPDKVQSLNVDAIIRGGLTRFTDQLGKLWCSLADYYIRSGHFEKARDVYE 280
Query 183 EAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQ 214
EA+ +V T+RD V+++YA FEE+++A ++
Sbjct 281 EAIRTVMTVRDFTQVFDSYAQFEESMIAAKME 312
> SPBC211.02c
Length=790
Score = 197 bits (502), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 98/270 (36%), Positives = 159/270 (58%), Gaps = 16/270 (5%)
Query 3 RCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCG 62
R L+ L +MP IW++ L+FL +Q +T++R TF++ALRAL VTQH+ +W Y + G
Sbjct 100 RSLILLHKMPVIWKLYLQFLMKQPNVTKIRCTFNSALRALPVTQHDDIWDMFTKYAEDIG 159
Query 63 VPETAVCL--YRRWIMIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRH 120
C+ YRR+I +EP + + L +G ++EAA + + +A ++ +
Sbjct 160 ---GLFCIHVYRRYIQVEPRAIENYIEILCKLGLWNEAARQYEDILNRPVFLSAKRKSNY 216
Query 121 ELWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDI 180
++WLE +LV HP+ ++ VE + R+GI RFSD LW LA +Y+R+G KAR
Sbjct 217 QIWLEFSELVVQHPDHTQNIDVEKVFRAGIKRFSDQAGKLWTYLAQYYIRIGDYEKARST 276
Query 181 YEEAVSSVSTIRDLAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAKD---IDFAIAR 237
+ E ++++ T+R+ ++++A+ FEE ++ A EA++ A D IDF +A
Sbjct 277 FYEGMNNIMTVRNFTIIFDAFVEFEEQWLS--------ARVEASSGNANDELSIDFHMAW 328
Query 238 LERLTXRRPLLISSCKLRQNPHNVHEWLAR 267
LE++ +RPL I+ LRQN +NV EWL R
Sbjct 329 LEKILDKRPLYINDVLLRQNINNVDEWLRR 358
> Hs7661952
Length=963
Score = 34.3 bits (77), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 46/97 (47%), Gaps = 10/97 (10%)
Query 120 HELWLE-LCKLVAAHPESIHSMRVEDILRSGIARFSDAVASLWCSLASHYV----RMGLL 174
ELWLE L ++ + + V D+ + + ++W + V + G L
Sbjct 146 EELWLEWLHDEISMAQDGLDREHVYDLFEKAVKDY--ICPNIWLEYGQYSVGGIGQKGGL 203
Query 175 AKARDIYEEAVSSVS--TIRDLAVVYEAYAAFEEAVV 209
K R ++E A+SSV + LA ++EAY FE A+V
Sbjct 204 EKVRSVFERALSSVGLHMTKGLA-LWEAYREFESAIV 239
> YDR416w
Length=859
Score = 32.3 bits (72), Expect = 1.1, Method: Composition-based stats.
Identities = 26/127 (20%), Positives = 52/127 (40%), Gaps = 2/127 (1%)
Query 143 EDILRSGIARFSDAVASLWCSLASHYVRMGLLAKARDIYEEAVSSVSTIRDLAVVYEAYA 202
E+ +R + D L SLA +Y+ G L D+ ++++ D +Y Y
Sbjct 288 EEFMRQMNGIYPDKWLFLILSLAKYYISRGRLDSCGDLLKKSLQQTLRYSDFDRIYNFYL 347
Query 203 AFEEAVVAQLLQEQQQASPE--AAAAAAKDIDFAIARLERLTXRRPLLISSCKLRQNPHN 260
FE+ +L + ++ + + + +A E L + ++ LRQ+ +
Sbjct 348 LFEQECSQFILGKLKENDSKFFNQKDWTEKLQAHMATFESLINLYDIYLNDVALRQDSNL 407
Query 261 VHEWLAR 267
V W+ R
Sbjct 408 VETWMKR 414
> At3g04240
Length=977
Score = 32.3 bits (72), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 57/142 (40%), Gaps = 21/142 (14%)
Query 129 LVAAHPESIHSMRVEDILRSGIARFSDAV------ASLWCSLASHYVRMGLLAKARDIYE 182
LV AH + M+ + ++ + + +AV A W +LA ++ G L +A Y+
Sbjct 188 LVDAHSNLGNLMKAQGLIHEAYSCYLEAVRIQPTFAIAWSNLAGLFMESGDLNRALQYYK 247
Query 183 EAVSSVSTIRD----LAVVYEAYAAFEEAVVAQLLQEQQQASPEAAAAAAK--------- 229
EAV D L VY+A EA++ Q Q P +A A
Sbjct 248 EAVKLKPAFPDAYLNLGNVYKALGRPTEAIMC--YQHALQMRPNSAMAFGNIASIYYEQG 305
Query 230 DIDFAIARLERLTXRRPLLISS 251
+D AI ++ R P + +
Sbjct 306 QLDLAIRHYKQALSRDPRFLEA 327
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 15/71 (21%)
Query 152 RFSDAVASLWCSLASHYVRMGLLAKARDIYEEAVS-------SVSTIRDL----AVVYEA 200
F+DA W +LAS Y+R G L++A ++A+S + S + +L +++EA
Sbjct 153 NFADA----WSNLASAYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIHEA 208
Query 201 YAAFEEAVVAQ 211
Y+ + EAV Q
Sbjct 209 YSCYLEAVRIQ 219
> At3g03580
Length=882
Score = 32.0 bits (71), Expect = 1.5, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 163 SLASHYVRMGLLAKARDIYEE-AVSSVSTIRDLAVVYEAYAAFEEAV 208
+L Y RMGLL +AR +++E V + + L Y ++ +EEA+
Sbjct 146 ALVDMYSRMGLLTRARQVFDEMPVRDLVSWNSLISGYSSHGYYEEAL 192
> 7290299
Length=702
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 48/106 (45%), Gaps = 9/106 (8%)
Query 19 LKFLQQQQLL---TRVRRTFDAALRALAVTQHELLWPEMMDYVKSCGVPETAVCLYRRWI 75
+KF + + LL R R F+ A++ + ELLW +D+ + G E A LY R +
Sbjct 478 MKFAELENLLGDTDRARAIFELAVQQPRLDMPELLWKAYIDFEVALGETELARQLYERLL 537
Query 76 MIEPERTDEFALYLSSIGRYDEAASHLAAVASDSTITTASGRTRHE 121
ERT +++ S +++ SH D+ + R +E
Sbjct 538 ----ERTQHVKVWM-SFAKFEMGLSH-GDSGPDAELNVQLARRIYE 577
> CE01471
Length=799
Score = 30.8 bits (68), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 41/83 (49%), Gaps = 16/83 (19%)
Query 1 GTRCLVHLSRMPEIWRMNLKFLQQQQLLTRVRRTFDAALRALAVTQHE-----------L 49
G+ CL HL +M I R+++K ++ + R+ DAA + ++TQ + L
Sbjct 408 GSACLYHLCKMKRIKRLSVK-----EVNNCIERSLDAAEQYRSMTQLQKNVWLTICNDYL 462
Query 50 LWPEMMDYVKSCGVPETAVCLYR 72
L + +D+ ++C V + L R
Sbjct 463 LHLDEIDFYRTCKVALDTMLLNR 485
> Hs17482900
Length=753
Score = 30.4 bits (67), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 32/82 (39%), Gaps = 0/82 (0%)
Query 99 ASHLAAVASDSTITTASGRTRHELWLELCKLVAAHPESIHSMRVEDILRSGIARFSDAVA 158
A L + S + +GR LW LC+L A + + + ++ G A F D
Sbjct 432 AQGLETLFSKAQELGGAGREDPGLWSRLCRLAGASSPAAYDEALAELHAHGPAAFVDYFE 491
Query 159 SLWCSLASHYVRMGLLAKARDI 180
W +VR ARD+
Sbjct 492 RNWEPRRDMWVRFRAFEAARDL 513
> Hs4505499
Length=920
Score = 30.4 bits (67), Expect = 3.7, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 39/78 (50%), Gaps = 10/78 (12%)
Query 153 FSDAVASLWCSLASHYVRMGLLAKARDIYEEAV----SSVSTIRDLAVVYEAYAAFEEAV 208
F DA +C+LA+ G +A+A D Y A+ + ++ +LA + EEAV
Sbjct 166 FPDA----YCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAV 221
Query 209 VAQLLQEQQQASPEAAAA 226
+L ++ + PE AAA
Sbjct 222 --RLYRKALEVFPEFAAA 237
> At4g08210
Length=686
Score = 30.4 bits (67), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 27/60 (45%), Gaps = 5/60 (8%)
Query 26 QLLTRVRRTFDAALRALAVTQHELLWPEMMDYVKSCGVPETAVCLYRRWIMIEPERTDEF 85
+LL+ + FD VT W M+ S G P A+ LYRR + E E +EF
Sbjct 54 RLLSDAHKVFDEMSERNIVT-----WTTMVSGYTSDGKPNKAIELYRRMLDSEEEAANEF 108
> CE28622
Length=744
Score = 29.6 bits (65), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 39/151 (25%), Positives = 68/151 (45%), Gaps = 21/151 (13%)
Query 8 LSRMPEIWRMNLKFLQQQQLL---TRVRRTFDAALRALAVTQHELLWPEMMDYVKSCGVP 64
L PE + +KF + + LL R R F A++ A+ ELLW +D+ +C
Sbjct 479 LESSPESSQTWIKFAELETLLGDTDRSRAVFTIAVQQPALDMPELLWKAYIDFEIACEEH 538
Query 65 ETAVCLYRRWIMIEPERTDEFALYLS------SIGRYDEAASHLAAVASDSTITTASGRT 118
E A LY + +RT+ +++S +IG ++ A A ++ ++ A
Sbjct 539 EKARDLYETLL----QRTNHIKVWISMAEFEQTIGNFEGARK--AFERANQSLENAEKEE 592
Query 119 R---HELWLELCKLVAAHPESIHSMRVEDIL 146
R E W E C+ + E++ RVE ++
Sbjct 593 RLMLLEAWKE-CETKSGDQEALK--RVETMM 620
Lambda K H
0.323 0.131 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5666887360
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40