bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4371_orf2
Length=148
Score E
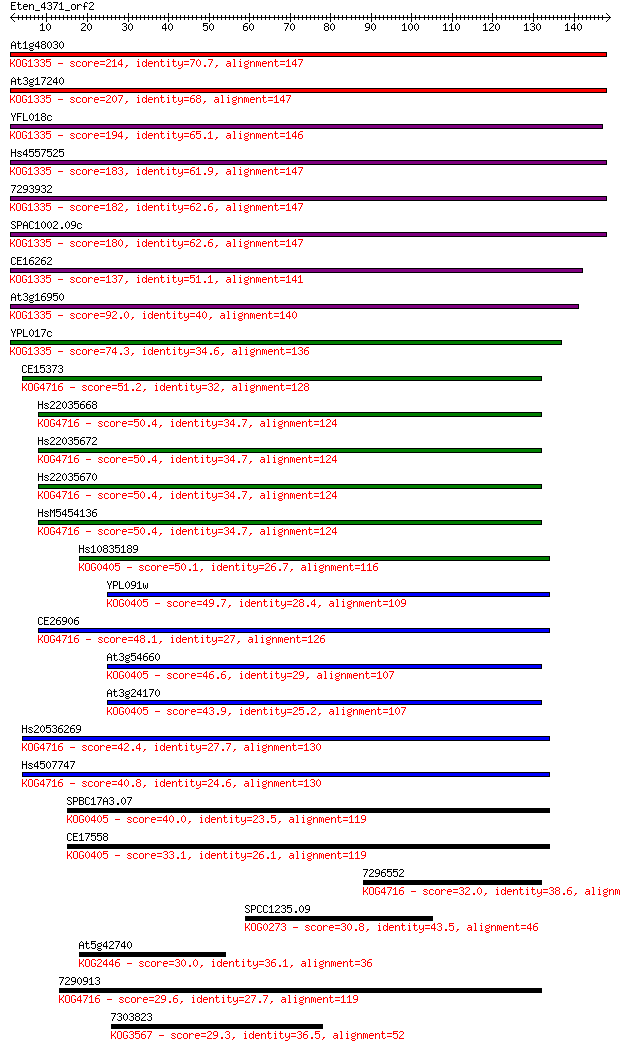
Sequences producing significant alignments: (Bits) Value
At1g48030 214 6e-56
At3g17240 207 7e-54
YFL018c 194 8e-50
Hs4557525 183 1e-46
7293932 182 2e-46
SPAC1002.09c 180 8e-46
CE16262 137 7e-33
At3g16950 92.0 4e-19
YPL017c 74.3 8e-14
CE15373 51.2 6e-07
Hs22035668 50.4 1e-06
Hs22035672 50.4 1e-06
Hs22035670 50.4 1e-06
HsM5454136 50.4 1e-06
Hs10835189 50.1 1e-06
YPL091w 49.7 2e-06
CE26906 48.1 6e-06
At3g54660 46.6 2e-05
At3g24170 43.9 1e-04
Hs20536269 42.4 3e-04
Hs4507747 40.8 0.001
SPBC17A3.07 40.0 0.002
CE17558 33.1 0.19
7296552 32.0 0.40
SPCC1235.09 30.8 1.1
At5g42740 30.0 1.7
7290913 29.6 2.2
7303823 29.3 3.2
> At1g48030
Length=507
Score = 214 bits (544), Expect = 6e-56, Method: Compositional matrix adjust.
Identities = 104/147 (70%), Positives = 121/147 (82%), Gaps = 1/147 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEE+G+ACVE +AGK G V +D +P V+YTHPE+A VGKTEEQLK EGV Y+ G F
Sbjct 362 AHKAEEDGVACVEFIAGK-HGHVDYDKVPGVVYTHPEVASVGKTEEQLKKEGVSYRVGKF 420
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF ANSRA+A +++GLVK+L KETDKILGV I+ PNAGELI EAVLA+ Y ASSED+
Sbjct 421 PFMANSRAKAIDNAEGLVKILADKETDKILGVHIMAPNAGELIHEAVLAINYDASSEDIA 480
Query 121 RVCHAHPTQSEALKEACMACFDKPIHI 147
RVCHAHPT SEALKEA MA +DKPIHI
Sbjct 481 RVCHAHPTMSEALKEAAMATYDKPIHI 507
> At3g17240
Length=507
Score = 207 bits (526), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 100/147 (68%), Positives = 119/147 (80%), Gaps = 1/147 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEE+G+ACVE +AGK G V +D +P V+YT+PE+A VGKTEEQLK EGV Y G F
Sbjct 362 AHKAEEDGVACVEFIAGK-HGHVDYDKVPGVVYTYPEVASVGKTEEQLKKEGVSYNVGKF 420
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF ANSRA+A ++G+VK+L KETDKILGV I+ PNAGELI EAVLA+ Y ASSED+
Sbjct 421 PFMANSRAKAIDTAEGMVKILADKETDKILGVHIMSPNAGELIHEAVLAINYDASSEDIA 480
Query 121 RVCHAHPTQSEALKEACMACFDKPIHI 147
RVCHAHPT SEA+KEA MA +DKPIH+
Sbjct 481 RVCHAHPTMSEAIKEAAMATYDKPIHM 507
> YFL018c
Length=499
Score = 194 bits (492), Expect = 8e-50, Method: Compositional matrix adjust.
Identities = 95/146 (65%), Positives = 115/146 (78%), Gaps = 1/146 (0%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAEEEGIA VE L G G V ++ IP+V+Y+HPE+A VGKTEEQLK G+ YK G F
Sbjct 354 AHKAEEEGIAAVEMLK-TGHGHVNYNNIPSVMYSHPEVAWVGKTEEQLKEAGIDYKIGKF 412
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRA+ D++G VK+L +T++ILG I+GPNAGE+IAEA LA+EYGAS+ED+
Sbjct 413 PFAANSRAKTNQDTEGFVKILIDSKTERILGAHIIGPNAGEMIAEAGLALEYGASAEDVA 472
Query 121 RVCHAHPTQSEALKEACMACFDKPIH 146
RVCHAHPT SEA KEA MA +DK IH
Sbjct 473 RVCHAHPTLSEAFKEANMAAYDKAIH 498
> Hs4557525
Length=509
Score = 183 bits (464), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 91/148 (61%), Positives = 114/148 (77%), Gaps = 2/148 (1%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGI CVE +AG G + ++ +P+VIYTHPE+A VGK+EEQLK EG+ YK G F
Sbjct 363 AHKAEDEGIICVEGMAG-GAVHIDYNCVPSVIYTHPEVAWVGKSEEQLKEEGIEYKVGKF 421
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PFAANSRA+ D+DG+VK+L K TD++LG ILGP AGE++ EA LA+EYGAS ED+
Sbjct 422 PFAANSRAKTNADTDGMVKILGQKSTDRVLGAHILGPGAGEMVNEAALALEYGASCEDIA 481
Query 121 RVCHAHPTQSEALKEACM-ACFDKPIHI 147
RVCHAHPT SEA +EA + A F K I+
Sbjct 482 RVCHAHPTLSEAFREANLAASFGKSINF 509
> 7293932
Length=504
Score = 182 bits (463), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 92/148 (62%), Positives = 114/148 (77%), Gaps = 2/148 (1%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EG+ +E + G G + ++ +P+V+YTHPE+A VGK+EEQLK EGV YK G F
Sbjct 358 AHKAEDEGLITIEGING-GHVHIDYNCVPSVVYTHPEVAWVGKSEEQLKQEGVAYKVGKF 416
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF ANSRA+ D+DG VKVL + TDKILG I+GP AGELI EAVLAMEYGA++ED+
Sbjct 417 PFLANSRAKTNNDTDGFVKVLADQATDKILGTHIIGPGAGELINEAVLAMEYGAAAEDVA 476
Query 121 RVCHAHPTQSEALKEACM-ACFDKPIHI 147
RVCHAHPT SEAL+EA + A F KPI+
Sbjct 477 RVCHAHPTCSEALREANVAAAFGKPINF 504
> SPAC1002.09c
Length=511
Score = 180 bits (457), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 92/148 (62%), Positives = 114/148 (77%), Gaps = 2/148 (1%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGIA VE +A KG G V ++ IPAV+YTHPE+A VG TE++ K G+ Y+ G F
Sbjct 365 AHKAEDEGIAAVEYIA-KGQGHVNYNCIPAVMYTHPEVAWVGITEQKAKESGIKYRIGTF 423
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF+ANSRA+ D+DGLVKV+ ETD++LGV ++GP AGELI EA LA+EYGAS+ED+
Sbjct 424 PFSANSRAKTNMDADGLVKVIVDAETDRLLGVHMIGPMAGELIGEATLALEYGASAEDVA 483
Query 121 RVCHAHPTQSEALKEACMACF-DKPIHI 147
RVCHAHPT SEA KEA MA + K IH
Sbjct 484 RVCHAHPTLSEATKEAMMAAWCGKSIHF 511
> CE16262
Length=464
Score = 137 bits (345), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 72/141 (51%), Positives = 86/141 (60%), Gaps = 32/141 (22%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGAF 60
AHKAE+EGI CVE +AG GV YK G F
Sbjct 348 AHKAEDEGILCVEGIAG--------------------------------GPGVAYKIGKF 375
Query 61 PFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLG 120
PF ANSRA+ D +G VKVL K+TD++LGV I+GPNAGE+IAEA LAMEYGAS+ED+
Sbjct 376 PFVANSRAKTNNDQEGFVKVLADKQTDRMLGVHIIGPNAGEMIAEATLAMEYGASAEDVA 435
Query 121 RVCHAHPTQSEALKEACMACF 141
RVCH HPT SEA +EA +A +
Sbjct 436 RVCHPHPTLSEAFREANLAAY 456
> At3g16950
Length=564
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 56/144 (38%), Positives = 81/144 (56%), Gaps = 5/144 (3%)
Query 1 AHKAEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLK----AEGVVYK 56
AH A +GI+ VE+++G+ D + H +IPA +THPE++ VG TE Q K EG
Sbjct 405 AHAASAQGISVVEQVSGR-DHVLNHLSIPAACFTHPEISMVGLTEPQAKEKGEKEGFKVS 463
Query 57 RGAFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASS 116
F AN++A A + +G+ K++ + +ILGV I G +A +LI EA A+ G
Sbjct 464 VVKTSFKANTKALAENEGEGIAKMIYRPDNGEILGVHIFGLHAADLIHEASNAIALGTRI 523
Query 117 EDLGRVCHAHPTQSEALKEACMAC 140
+D+ HAHPT SE L E A
Sbjct 524 QDIKLAVHAHPTLSEVLDELFKAA 547
> YPL017c
Length=499
Score = 74.3 bits (181), Expect = 8e-14, Method: Composition-based stats.
Identities = 47/143 (32%), Positives = 74/143 (51%), Gaps = 7/143 (4%)
Query 1 AHKAEEEGIACVERLAGKG-DGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGVVYKRGA 59
A KAEE+ I ++ + G DGT P V+Y P++ VG TEE L + Y++G
Sbjct 340 ALKAEEQAIRAIQSIGCTGSDGTSNCGFPPNVLYCQPQIGWVGYTEEGLAKARIPYQKGR 399
Query 60 FPFAANSRARA------TGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYG 113
F+ N R +KVL KILGV ++ +A EL+++A +A+ G
Sbjct 400 VLFSQNVRYNTLLPREENTTVSPFIKVLIDSRDMKILGVHMINDDANELLSQASMAVSLG 459
Query 114 ASSEDLGRVCHAHPTQSEALKEA 136
++ D+ +V HP+ SE+ K+A
Sbjct 460 LTAHDVCKVPFPHPSLSESFKQA 482
> CE15373
Length=503
Score = 51.2 bits (121), Expect = 6e-07, Method: Composition-based stats.
Identities = 41/133 (30%), Positives = 57/133 (42%), Gaps = 5/133 (3%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ-LKAEGV----VYKRG 58
A + G +RL VR D + ++T EL+ VG TEE+ ++ G V+
Sbjct 349 AIQSGKLLADRLFSNSKQIVRFDGVATTVFTPLELSTVGLTEEEAIQKHGEDSIEVFHSH 408
Query 59 AFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSED 118
PF + V T E+ KILG+ +GPNA E+I +A G S D
Sbjct 409 FTPFEYVVPQNKDSGFCYVKAVCTRDESQKILGLHFVGPNAAEVIQGYAVAFRVGISMSD 468
Query 119 LGRVCHAHPTQSE 131
L HP SE
Sbjct 469 LQNTIAIHPCSSE 481
> Hs22035668
Length=428
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/130 (33%), Positives = 61/130 (46%), Gaps = 8/130 (6%)
Query 8 GIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA----EGV-VYKRGAFPF 62
G V+RL G + +D +P ++T E VG +EE+ A E V VY P
Sbjct 279 GRLLVQRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPL 338
Query 63 AANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGASSEDLGR 121
R S VK++ +E + +LG+ LGPNAGE+ L ++ GAS + R
Sbjct 339 EFTVAGRDA--SQCYVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMR 396
Query 122 VCHAHPTQSE 131
HPT SE
Sbjct 397 TVGIHPTCSE 406
> Hs22035672
Length=524
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/130 (33%), Positives = 61/130 (46%), Gaps = 8/130 (6%)
Query 8 GIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA----EGV-VYKRGAFPF 62
G V+RL G + +D +P ++T E VG +EE+ A E V VY P
Sbjct 375 GRLLVQRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPL 434
Query 63 AANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGASSEDLGR 121
R S VK++ +E + +LG+ LGPNAGE+ L ++ GAS + R
Sbjct 435 EFTVAGRDA--SQCYVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMR 492
Query 122 VCHAHPTQSE 131
HPT SE
Sbjct 493 TVGIHPTCSE 502
> Hs22035670
Length=494
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/130 (33%), Positives = 61/130 (46%), Gaps = 8/130 (6%)
Query 8 GIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA----EGV-VYKRGAFPF 62
G V+RL G + +D +P ++T E VG +EE+ A E V VY P
Sbjct 345 GRLLVQRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPL 404
Query 63 AANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGASSEDLGR 121
R S VK++ +E + +LG+ LGPNAGE+ L ++ GAS + R
Sbjct 405 EFTVAGRDA--SQCYVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMR 462
Query 122 VCHAHPTQSE 131
HPT SE
Sbjct 463 TVGIHPTCSE 472
> HsM5454136
Length=524
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/130 (33%), Positives = 61/130 (46%), Gaps = 8/130 (6%)
Query 8 GIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKA----EGV-VYKRGAFPF 62
G V+RL G + +D +P ++T E VG +EE+ A E V VY P
Sbjct 375 GRLLVQRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPL 434
Query 63 AANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGASSEDLGR 121
R S VK++ +E + +LG+ LGPNAGE+ L ++ GAS + R
Sbjct 435 EFTVAGRDA--SQCYVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMR 492
Query 122 VCHAHPTQSE 131
HPT SE
Sbjct 493 TVGIHPTCSE 502
> Hs10835189
Length=479
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 58/118 (49%), Gaps = 2/118 (1%)
Query 18 KGDGTVRHDTIPAVIYTHPELAGVGKTE-EQLKAEGV-VYKRGAFPFAANSRARATGDSD 75
K D + ++ IP V+++HP + VG TE E + G+ K + F A +
Sbjct 358 KEDSKLDYNNIPTVVFSHPPIGTVGLTEDEAIHKYGIENVKTYSTSFTPMYHAVTKRKTK 417
Query 76 GLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
++K++ + +K++G+ + G E++ +A++ GA+ D HPT SE L
Sbjct 418 CVMKMVCANKEEKVVGIHMQGLGCDEMLQGFAVAVKMGATKADFDNTVAIHPTSSEEL 475
> YPL091w
Length=483
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 31/114 (27%), Positives = 58/114 (50%), Gaps = 8/114 (7%)
Query 25 HDTIPAVIYTHPELAGVGKTE----EQLKAEGV-VYKRGAFPFAANSRARATGDSDGLVK 79
++ +P+VI++HPE +G +E E+ E + VY F A A + S K
Sbjct 369 YENVPSVIFSHPEAGSIGISEKEAIEKYGKENIKVYNS---KFTAMYYAMLSEKSPTRYK 425
Query 80 VLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSEAL 133
++ +K++G+ I+G ++ E++ +A++ GA+ D HPT +E L
Sbjct 426 IVCAGPNEKVVGLHIVGDSSAEILQGFGVAIKMGATKADFDNCVAIHPTSAEEL 479
> CE26906
Length=665
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 34/131 (25%), Positives = 53/131 (40%), Gaps = 5/131 (3%)
Query 8 GIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQL-----KAEGVVYKRGAFPF 62
G + R+ + +D IP ++T E G +EE K ++Y P
Sbjct 516 GRVLMRRIFDGANELTEYDQIPTTVFTPLEYGCCGLSEEDAMMKYGKDNIIIYHNVFNPL 575
Query 63 AANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRV 122
R D L + E +K++G IL PNAGE+ +A++ A D R+
Sbjct 576 EYTISERMDKDHCYLKMICLRNEEEKVVGFHILTPNAGEVTQGFGIALKLAAKKADFDRL 635
Query 123 CHAHPTQSEAL 133
HPT +E
Sbjct 636 IGIHPTVAENF 646
> At3g54660
Length=565
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 54/111 (48%), Gaps = 8/111 (7%)
Query 25 HDTIPAVIYTHPELAGVGKTEEQL---KAEGVVYKRGAFPFAANSRARATGDSDG-LVKV 80
+ +P +++ P + VG TEEQ + VY P +A +G D +K+
Sbjct 428 YRAVPCAVFSQPPIGTVGLTEEQAIEQYGDVDVYTSNFRPL----KATLSGLPDRVFMKL 483
Query 81 LTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
+ C T+K+LGV + G ++ E+I +A++ G + D HPT +E
Sbjct 484 IVCANTNKVLGVHMCGEDSPEIIQGFGVAVKAGLTKADFDATVGVHPTAAE 534
> At3g24170
Length=499
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/111 (24%), Positives = 53/111 (47%), Gaps = 7/111 (6%)
Query 25 HDTIPAVIYTHPELAGVGKTEE----QLKAEGVVYKRGAFPFAANSRARATGDSDGLVKV 80
+ + ++ P LA VG +EE Q + +V+ G P R L+K+
Sbjct 370 YSNVACAVFCIPPLAVVGLSEEEAVEQATGDILVFTSGFNPMKNTISGR---QEKTLMKL 426
Query 81 LTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
+ +++DK++G + GP+A E++ +A++ GA+ HP+ +E
Sbjct 427 IVDEKSDKVIGASMCGPDAAEIMQGIAIALKCGATKAQFDSTVGIHPSSAE 477
> Hs20536269
Length=459
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 36/136 (26%), Positives = 59/136 (43%), Gaps = 8/136 (5%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ----LKAEGV-VYKRG 58
A + G +RL G + +P ++T E G +EE+ K E + +Y
Sbjct 308 AIQSGKLLAQRLFGASLEKCDYINVPTTVFTPLEYGCCGLSEEKAIEVYKKENLEIYHTL 367
Query 59 AFPFAANSRARATGDSDGLVKVLTCK-ETDKILGVWILGPNAGELIAEAVLAMEYGASSE 117
+P R ++ K++ K + D+++G ILGPNAGE+ AM+ G + +
Sbjct 368 FWPLEWTVAGRE--NNTCYAKIICNKFDHDRVIGFHILGPNAGEVTQGFAAAMKCGLTKQ 425
Query 118 DLGRVCHAHPTQSEAL 133
L HPT E
Sbjct 426 LLDDTIGIHPTCGEVF 441
> Hs4507747
Length=497
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/137 (23%), Positives = 59/137 (43%), Gaps = 10/137 (7%)
Query 4 AEEEGIACVERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQ----LKAEGV-VYKRG 58
A + G +RL ++ +P ++T E G +EE+ E + VY
Sbjct 346 AIQAGRLLAQRLYAGSTVKCDYENVPTTVFTPLEYGACGLSEEKAVEKFGEENIEVYHSY 405
Query 59 AFPFAANSRARATGDSDGLVKVLTC--KETDKILGVWILGPNAGELIAEAVLAMEYGASS 116
+P +R D++ + C K+ ++++G +LGPNAGE+ A++ G +
Sbjct 406 FWPLEWTIPSR---DNNKCYAKIICNTKDNERVVGFHVLGPNAGEVTQGFAAALKCGLTK 462
Query 117 EDLGRVCHAHPTQSEAL 133
+ L HP +E
Sbjct 463 KQLDSTIGIHPVCAEVF 479
> SPBC17A3.07
Length=464
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/124 (22%), Positives = 56/124 (45%), Gaps = 6/124 (4%)
Query 15 LAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQL-----KAEGVVYKRGAFPFAANSRAR 69
G D + ++ +P+V++ HPE +G TE++ +++ VY F S
Sbjct 338 FGGIKDAHLDYEEVPSVVFAHPEAGTIGLTEQEAIDKYGESQIKVYNT-KFNGLNYSMVE 396
Query 70 ATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQ 129
K++ K++G+ ++G + E++ +A++ GA+ D HPT
Sbjct 397 QEDKVPTTYKLVCAGPLQKVVGLHLVGDFSAEILQGFGVAIKMGATKSDFDSCVAIHPTS 456
Query 130 SEAL 133
+E L
Sbjct 457 AEEL 460
> CE17558
Length=473
Score = 33.1 bits (74), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 31/126 (24%), Positives = 58/126 (46%), Gaps = 12/126 (9%)
Query 15 LAGKGDGTVRHDTIPAVIYTHPELAGVGKTEEQL-----KAEGVVYKRGAFP--FAANSR 67
G+ D + ++ I V+++HP + VG TE + K E +YK P FA
Sbjct 343 FNGETDNKLTYENIATVVFSHPLIGTVGLTEAEAVEKYGKDEVTLYKSRFNPMLFAVTKH 402
Query 68 ARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHP 127
+K++ + +K++GV + G + E++ +A+ GA+ + + HP
Sbjct 403 KEKAA-----MKLVCVGKDEKVVGVHVFGVGSDEMLQGFAVAVTMGATKKQFDQTVAIHP 457
Query 128 TQSEAL 133
T +E L
Sbjct 458 TSAEEL 463
> 7296552
Length=516
Score = 32.0 bits (71), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 88 KILGVWILGPNAGELIAEAVLAMEYGASSEDLGRVCHAHPTQSE 131
KILG+ +GP AGE+I A++ G + + L HPT +E
Sbjct 451 KILGLHYIGPVAGEVIQGFAAALKTGLTVKTLLNTVGIHPTTAE 494
> SPCC1235.09
Length=564
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 3/46 (6%)
Query 59 AFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIA 104
A F+ N R ATGD+ G V + +CK T K+ LG + ELIA
Sbjct 480 ALSFSHNGRYLATGDTSGGVCIWSCK-TAKLFKE--LGSDNSELIA 522
> At5g42740
Length=560
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 18 KGDGTVRHDTIPAVIYTHPELAGVGKTEEQLKAEGV 53
KG+ HD + + + P+ GKT EQL+ E V
Sbjct 421 KGEVVSNHDELMSNFFAQPDALAYGKTPEQLQKENV 456
> 7290913
Length=491
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 33/129 (25%), Positives = 53/129 (41%), Gaps = 16/129 (12%)
Query 13 ERLAGKGDGTVRHDTIPAVIYTHPELAGVGKTEE----QLKAEGV-----VYKRGAFPFA 63
RL G + + + ++T E A VG +EE Q A+ + YK F F
Sbjct 347 RRLYGGSTQRMDYKDVATTVFTPLEYACVGLSEEDAVKQFGADEIEVFHGYYKPTEF-FI 405
Query 64 ANSRARATGDSDGLVKVLTCKETD-KILGVWILGPNAGELIAEAVLAMEYGASSEDLGRV 122
R +K + + D ++ G+ +GP AGE+I A++ G + L
Sbjct 406 PQKSVRYC-----YLKAVAERHGDQRVYGLHYIGPVAGEVIQGFAAALKSGLTINTLINT 460
Query 123 CHAHPTQSE 131
HPT +E
Sbjct 461 VGIHPTTAE 469
> 7303823
Length=541
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 22/62 (35%), Gaps = 10/62 (16%)
Query 26 DTIPAVIYTHPELAGVGKTEEQLKAEGVVY----------KRGAFPFAANSRARATGDSD 75
D +PAV HP + L G + KRG PF A R A SD
Sbjct 444 DQVPAVAVHHPSGKAILVASLMLLFAGSTFALALIFARRRKRGCLPFGARGRRHAWEKSD 503
Query 76 GL 77
G
Sbjct 504 GF 505
Lambda K H
0.316 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40