bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4330_orf3
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
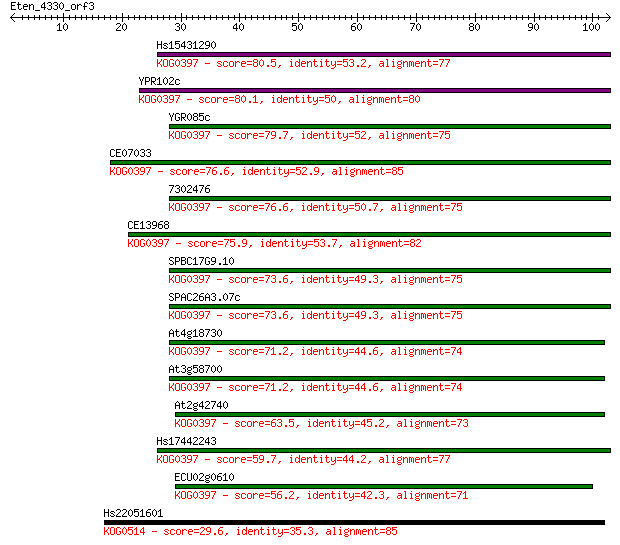
Hs15431290 80.5 6e-16
YPR102c 80.1 1e-15
YGR085c 79.7 1e-15
CE07033 76.6 9e-15
7302476 76.6 9e-15
CE13968 75.9 2e-14
SPBC17G9.10 73.6 1e-13
SPAC26A3.07c 73.6 1e-13
At4g18730 71.2 4e-13
At3g58700 71.2 5e-13
At2g42740 63.5 9e-11
Hs17442243 59.7 1e-09
ECU02g0610 56.2 2e-08
Hs22051601 29.6 1.4
> Hs15431290
Length=178
Score = 80.5 bits (197), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 41/77 (53%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 26 ERPTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKN 85
E P R+ R K N GE+GDR TRAA+ EQ TGQ P ++AR T R+ RRNEK
Sbjct 9 ENPMRELRIRKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTVRSFGIRRNEKI 68
Query 86 ARHETARGKKAEETEEK 102
A H T RG KAEE EK
Sbjct 69 AVHCTVRGAKAEEILEK 85
> YPR102c
Length=174
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 40/80 (50%), Positives = 49/80 (61%), Gaps = 0/80 (0%)
Query 23 ATEERPTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRN 82
A + P R + EK N GE+GDR TRA++ EQ +GQ P Q++AR T RT RRN
Sbjct 3 AKAQNPMRDLKIEKLVLNISVGESGDRLTRASKVLEQLSGQTPVQSKARYTVRTFGIRRN 62
Query 83 EKNARHETARGKKAEETEEK 102
EK A H T RG KAEE E+
Sbjct 63 EKIAVHVTVRGPKAEEILER 82
> YGR085c
Length=174
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/75 (52%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 28 PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKNAR 87
P R + EK N GE+GDR TRA++ EQ +GQ P Q++AR T RT RRNEK A
Sbjct 8 PMRDLKIEKLVLNISVGESGDRLTRASKVLEQLSGQTPVQSKARYTVRTFGIRRNEKIAV 67
Query 88 HETARGKKAEETEEK 102
H T RG KAEE E+
Sbjct 68 HVTVRGPKAEEILER 82
> CE07033
Length=196
Score = 76.6 bits (187), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 45/91 (49%), Positives = 52/91 (57%), Gaps = 6/91 (6%)
Query 18 GKKEKATEER------PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRAR 71
G EK TE R R+ + +K N GE+GDR TRAA+ EQ TGQ P ++AR
Sbjct 2 GDIEKQTEIREKKARNVMRELKIQKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKAR 61
Query 72 KTSRTDSKRRNEKNARHETARGKKAEETEEK 102
T RT RRNEK A H T RG KAEE EK
Sbjct 62 YTVRTFGIRRNEKIAVHCTVRGPKAEEILEK 92
> 7302476
Length=184
Score = 76.6 bits (187), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 38/75 (50%), Positives = 45/75 (60%), Gaps = 0/75 (0%)
Query 28 PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKNAR 87
P R K N GE+GDR TRAA+ EQ TGQ+P ++AR T R+ RRNEK A
Sbjct 16 PMRDLHIRKLCLNICVGESGDRLTRAAKVLEQLTGQQPVFSKARYTVRSFGIRRNEKIAV 75
Query 88 HETARGKKAEETEEK 102
H T RG KAEE E+
Sbjct 76 HCTVRGAKAEEILER 90
> CE13968
Length=196
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/88 (50%), Positives = 51/88 (57%), Gaps = 6/88 (6%)
Query 21 EKATEER------PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTS 74
EK TE R R+ + +K N GE+GDR TRAA+ EQ TGQ P ++AR T
Sbjct 5 EKQTEIREKKGRNVMRELKIQKLCLNICVGESGDRLTRAAKVLEQLTGQTPVFSKARYTV 64
Query 75 RTDSKRRNEKNARHETARGKKAEETEEK 102
RT RRNEK A H T RG KAEE EK
Sbjct 65 RTFGIRRNEKIAVHCTVRGPKAEEILEK 92
> SPBC17G9.10
Length=174
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/75 (49%), Positives = 45/75 (60%), Gaps = 0/75 (0%)
Query 28 PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKNAR 87
P ++ R K N GE+GDR TRAA+ EQ +GQ P ++AR T R RRNEK A
Sbjct 8 PMKELRISKLVLNISLGESGDRLTRAAKVLEQLSGQTPVFSKARYTIRRFGIRRNEKIAC 67
Query 88 HETARGKKAEETEEK 102
H T RG KAEE E+
Sbjct 68 HVTVRGPKAEEILER 82
> SPAC26A3.07c
Length=174
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/75 (49%), Positives = 45/75 (60%), Gaps = 0/75 (0%)
Query 28 PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKNAR 87
P ++ R K N GE+GDR TRAA+ EQ +GQ P ++AR T R RRNEK A
Sbjct 8 PMKELRISKLVLNISLGESGDRLTRAAKVLEQLSGQTPVFSKARYTIRRFGIRRNEKIAC 67
Query 88 HETARGKKAEETEEK 102
H T RG KAEE E+
Sbjct 68 HVTVRGPKAEEILER 82
> At4g18730
Length=184
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 28 PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKNAR 87
P R + +K N GE+GDR TRA++ EQ +GQ P ++AR T R+ RRNEK A
Sbjct 12 PMRDIKVQKLVLNISVGESGDRLTRASKVLEQLSGQTPVFSKARYTVRSFGIRRNEKIAC 71
Query 88 HETARGKKAEETEE 101
+ T RG+KA + E
Sbjct 72 YVTVRGEKAMQLLE 85
> At3g58700
Length=182
Score = 71.2 bits (173), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 28 PTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKNAR 87
P R + +K N GE+GDR TRA++ EQ +GQ P ++AR T R+ RRNEK A
Sbjct 10 PMRDIKVQKLVLNISVGESGDRLTRASKVLEQLSGQTPVFSKARYTVRSFGIRRNEKIAC 69
Query 88 HETARGKKAEETEE 101
+ T RG+KA + E
Sbjct 70 YVTVRGEKAMQLLE 83
> At2g42740
Length=182
Score = 63.5 bits (153), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 45/83 (54%), Gaps = 10/83 (12%)
Query 29 TRKNRSEKRTTNNGAGETGDRPTRAAREP----------EQRTGQRPAQTRARKTSRTDS 78
R + +K N GE+GDR TRA++ P EQ +GQ P ++AR T R+
Sbjct 1 MRDIKVQKLVLNISVGESGDRLTRASKVPPVFYPFSVVLEQLSGQTPVFSKARYTVRSFG 60
Query 79 KRRNEKNARHETARGKKAEETEE 101
RRNEK A + T RG+KA + E
Sbjct 61 IRRNEKIACYVTVRGEKAMQLLE 83
> Hs17442243
Length=171
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 41/77 (53%), Gaps = 6/77 (7%)
Query 26 ERPTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKN 85
E P +K K N E+GDR TRAA+ EQ TGQ+ T R+ +RNEK
Sbjct 8 ENPMQKLHICKLCLNICVEESGDRLTRAAKALEQLTGQK------LDTIRSSGMQRNEKI 61
Query 86 ARHETARGKKAEETEEK 102
A H T +G KAEE EK
Sbjct 62 AVHCTVQGSKAEEILEK 78
> ECU02g0610
Length=165
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 40/71 (56%), Gaps = 0/71 (0%)
Query 29 TRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRTDSKRRNEKNARH 88
R+ +K + + A E+G + RAA+ EQ TGQ+P ++AR T R RRNEK A H
Sbjct 1 MREISIKKLSIHICARESGAKLDRAAKVLEQLTGQKPVTSKARMTIRNFGIRRNEKIAVH 60
Query 89 ETARGKKAEET 99
GKKA E
Sbjct 61 VNVSGKKAYEL 71
> Hs22051601
Length=454
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 43/86 (50%), Gaps = 7/86 (8%)
Query 17 DGKKEKATEERPTRKNRSEKRTTNNGAGETGDRPTRAAREPEQRTGQRPAQTRARKTSRT 76
+G E ++E+ T +N S+ +T N A E +R A P+ R PA T A KTSR
Sbjct 118 NGGYESSSEDSSTAENISDNDSTENEAPEPRERVPSVAEAPQLR----PAGTAAAKTSRQ 173
Query 77 DSKRRNEKNARHE-TARGKKAEETEE 101
+ + E ++H TA G TEE
Sbjct 174 ECQLSRE--SQHIPTAEGASGSNTEE 197
Lambda K H
0.297 0.114 0.301
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181107380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40