bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
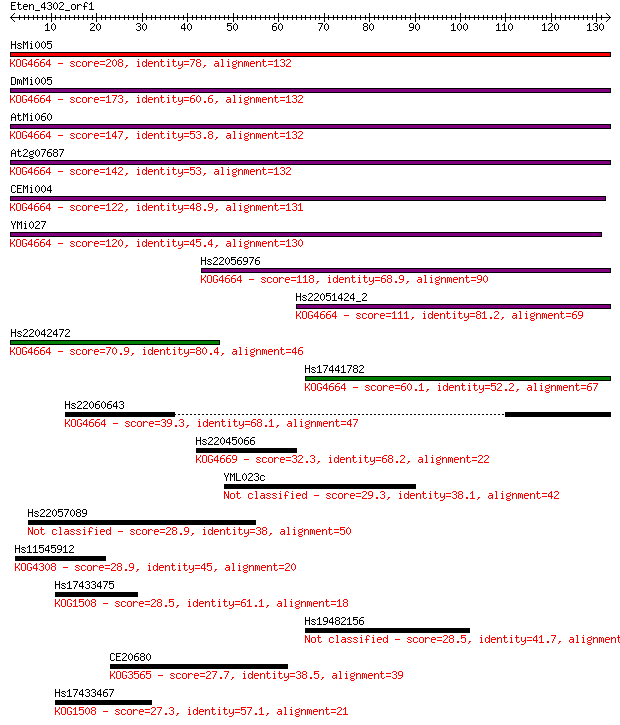
Query= Eten_4302_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
HsMi005 208 2e-54
DmMi005 173 9e-44
AtMi060 147 6e-36
At2g07687 142 1e-34
CEMi004 122 1e-28
YMi027 120 1e-27
Hs22056976 118 2e-27
Hs22051424_2 111 4e-25
Hs22042472 70.9 7e-13
Hs17441782 60.1 1e-09
Hs22060643 39.3 0.002
Hs22045066 32.3 0.27
YML023c 29.3 2.3
Hs22057089 28.9 2.5
Hs11545912 28.9 2.7
Hs17433475 28.5 3.4
Hs19482156 28.5 4.0
CE20680 27.7 5.5
Hs17433467 27.3 8.2
> HsMi005
Length=260
Score = 208 bits (529), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 103/132 (78%), Positives = 112/132 (84%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
PTP+LGG WPPTGI PLNPLEVPLLNTSVLLASGVSIT A HSL+E R ++QAL I
Sbjct 108 PTPQLGGHWPPTGITPLNPLEVPLLNTSVLLASGVSITWAHHSLMENNRNQMIQALLITI 167
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
LG+YFTLLQASEY+E+PFTISDG+YGSTFFVATGFHGLHVIIGS FL +CF QL FHF
Sbjct 168 LLGLYFTLLQASEYFESPFTISDGIYGSTFFVATGFHGLHVIIGSTFLTICFIRQLMFHF 227
Query 121 TSNPHFGFETAA 132
TS HFGFE AA
Sbjct 228 TSKHHFGFEAAA 239
> DmMi005
Length=262
Score = 173 bits (438), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 80/132 (60%), Positives = 98/132 (74%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P ELG WPP GI NP ++PLLNT++LLASGV++T A HSL+E Q LFF +
Sbjct 109 PAIELGASWPPMGIISFNPFQIPLLNTAILLASGVTVTWAHHSLMENNHSQTTQGLFFTV 168
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
LG+YFT+LQA EY EAPFTI+D +YGSTFF+ATGFHG+HV+IG+ FL+VC L HF
Sbjct 169 LLGIYFTILQAYEYIEAPFTIADSIYGSTFFMATGFHGIHVLIGTTFLLVCLLRHLNNHF 228
Query 121 TSNPHFGFETAA 132
+ N HFGFE AA
Sbjct 229 SKNHHFGFEAAA 240
> AtMi060
Length=265
Score = 147 bits (371), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 71/132 (53%), Positives = 89/132 (67%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P E+GG WPP GI L+P E+P LNT +LL+SG ++T A H+++ G K + AL +
Sbjct 111 PAVEIGGIWPPKGIEVLDPWEIPFLNTLILLSSGAAVTWAHHAILAGKEKRAVYALVATV 170
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L + FT Q EYY+APFTISD +YGSTFF+ATGFHG HVIIG+ FLI+C Q H
Sbjct 171 LLALVFTGFQGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHL 230
Query 121 TSNPHFGFETAA 132
T H GFE AA
Sbjct 231 TKEHHVGFEAAA 242
> At2g07687
Length=265
Score = 142 bits (359), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 70/132 (53%), Positives = 88/132 (66%), Gaps = 0/132 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P E+GG WPP GI L+P E+P LNT +L +SG ++T A H+++ G K + AL +
Sbjct 111 PAVEIGGIWPPKGIEVLDPWEIPFLNTPILPSSGAAVTWAHHAILAGKEKRAVYALVATV 170
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L + FT Q EYY+APFTISD +YGSTFF+ATGFHG HVIIG+ FLI+C Q H
Sbjct 171 LLALVFTGFQGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHL 230
Query 121 TSNPHFGFETAA 132
T H GFE AA
Sbjct 231 TKEHHVGFEAAA 242
> CEMi004
Length=255
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 64/131 (48%), Positives = 82/131 (62%), Gaps = 2/131 (1%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P ELG W P G+H +NP VPLLNT +LL+SGV++T A HSL+ K ++
Sbjct 104 PVHELGETWSPFGMHLVNPFGVPLLNTIILLSSGVTVTWAHHSLLS--NKSCTNSMILTC 161
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L YFT +Q EY EA F+I+DGV+GS F+++TGFHG+HV+ G FL F LK HF
Sbjct 162 LLAAYFTGIQLMEYMEASFSIADGVFGSIFYLSTGFHGIHVLCGGLFLAFNFLRLLKNHF 221
Query 121 TSNPHFGFETA 131
N H G E A
Sbjct 222 NYNHHLGLEFA 232
> YMi027
Length=269
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 59/130 (45%), Positives = 77/130 (59%), Gaps = 0/130 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPI 60
P LG CWPP GI + P E+PLLNT +LL+SG ++T + H+LI G R L L
Sbjct 116 PDVTLGACWPPVGIEAVQPTELPLLNTIILLSSGATVTYSHHALIAGNRNKALSGLLITF 175
Query 61 TLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHF 120
L V F Q EY A FTISDGVYGS F+ TG H LH+++ + L V ++ +H
Sbjct 176 WLIVIFVTCQYIEYTNAAFTISDGVYGSVFYAGTGLHFLHMVMLAAMLGVNYWRMRNYHL 235
Query 121 TSNPHFGFET 130
T+ H G+ET
Sbjct 236 TAGHHVGYET 245
> Hs22056976
Length=118
Score = 118 bits (296), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 62/90 (68%), Positives = 68/90 (75%), Gaps = 0/90 (0%)
Query 43 SLIEGGRKHILQALFFPITLGVYFTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVI 102
SLIEG RK ILQAL ITLG+Y TLL+ SEY+EAPF IS G+YGSTFF ATGF GL+VI
Sbjct 29 SLIEGSRKQILQALAITITLGIYATLLRVSEYFEAPFPISHGLYGSTFFTATGFPGLYVI 88
Query 103 IGSPFLIVCFFPQLKFHFTSNPHFGFETAA 132
I S L + QLKFHFTSN HFGFE A
Sbjct 89 IESTLLTIRLLCQLKFHFTSNHHFGFEATA 118
> Hs22051424_2
Length=77
Score = 111 bits (277), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 56/70 (80%), Positives = 59/70 (84%), Gaps = 1/70 (1%)
Query 64 VYFTLLQASEYYEAPFTISD-GVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHFTS 122
VYFTLLQASEY+E PFTISD G+YGSTFFVATGFHGLHVIIGS FL +C QL FHFTS
Sbjct 7 VYFTLLQASEYFETPFTISDDGIYGSTFFVATGFHGLHVIIGSTFLTICLIRQLTFHFTS 66
Query 123 NPHFGFETAA 132
HFGFE AA
Sbjct 67 KHHFGFEAAA 76
> Hs22042472
Length=144
Score = 70.9 bits (172), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 37/46 (80%), Positives = 38/46 (82%), Gaps = 0/46 (0%)
Query 1 PTPELGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIE 46
PTPELGGCWPPT I P + LEVP LNTSVLLASGVSIT A SLIE
Sbjct 54 PTPELGGCWPPTSISPPSLLEVPFLNTSVLLASGVSITWAHQSLIE 99
> Hs17441782
Length=102
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 66 FTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHVIIGSPFLIVCFFPQLKFHFTSNPH 125
FT+ Y I + FF A GFHGLHVII S FL +C LKFHFTSN H
Sbjct 37 FTIALIQAYVLTLLGIILFIISEIFFFA-GFHGLHVIIRSTFLTICLLRPLKFHFTSNHH 95
Query 126 FGFETAA 132
FGFE AA
Sbjct 96 FGFEAAA 102
> Hs22060643
Length=130
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 13 GIHPLNPLEVPLLNTSVLLASGVS 36
GI PL+PLEVPLLN SVLLASG+
Sbjct 70 GISPLDPLEVPLLNPSVLLASGLQ 93
Score = 32.3 bits (72), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 110 VCFFPQLKFHFTSNPHFGFETAA 132
VC QL +HFTS+ F FE AA
Sbjct 108 VCLLRQLNYHFTSSHQFAFEAAA 130
> Hs22045066
Length=300
Score = 32.3 bits (72), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 42 HSLIEGGRKHILQALFFPITLG 63
H LIEG RK ILQAL ITL
Sbjct 237 HCLIEGSRKQILQALSITITLA 258
> YML023c
Length=556
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 48 GRKHILQALFFPITLGVYFTLLQASEYYEAPFTISDGVYGST 89
G+ H L + F I L V FTL SEYY+ P ++ Y ++
Sbjct 56 GKSHNLAVIPFDIIL-VLFTLSTLSEYYKEPILRANDPYNTS 96
> Hs22057089
Length=205
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Query 5 LGGCWPPTGIHPLNPLEVPLLNTSVLLASGVSITGAPHSLIEGGRKHILQ 54
L GC P +GI VPLL + +A+G + TG L+ GR LQ
Sbjct 113 LTGCIPLSGISA-----VPLLRNASKVAAGGAGTGIQRPLVRRGRLKALQ 157
> Hs11545912
Length=1040
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 9/20 (45%), Positives = 13/20 (65%), Gaps = 0/20 (0%)
Query 2 TPELGGCWPPTGIHPLNPLE 21
+P+L GCW P +HP L+
Sbjct 116 SPKLHGCWDPHSLHPARDLQ 135
> Hs17433475
Length=420
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 11/18 (61%), Positives = 13/18 (72%), Gaps = 0/18 (0%)
Query 11 PTGIHPLNPLEVPLLNTS 28
PTG HP NP+ + LNTS
Sbjct 342 PTGSHPANPMRLGSLNTS 359
> Hs19482156
Length=519
Score = 28.5 bits (62), Expect = 4.0, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 4/36 (11%)
Query 66 FTLLQASEYYEAPFTISDGVYGSTFFVATGFHGLHV 101
F LL + EA F + +YGSTF FHG H+
Sbjct 447 FLLLSSPPAKEARFRTAKKLYGSTF----AFHGSHI 478
> CE20680
Length=1059
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 23 PLLNTSVLLASGVSITGAPHSLIEGGRKHILQALFFPIT 61
PLL+T + A+ SI+ AP ++ RK L+ PIT
Sbjct 899 PLLSTKYVSAASCSISPAPRTIDIKERKDSRDLLYAPIT 937
> Hs17433467
Length=368
Score = 27.3 bits (59), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 14/21 (66%), Gaps = 0/21 (0%)
Query 11 PTGIHPLNPLEVPLLNTSVLL 31
PTG HP NP+ + LNTS L
Sbjct 290 PTGSHPTNPMRLGSLNTSEAL 310
Lambda K H
0.326 0.145 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40