bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4296_orf2
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
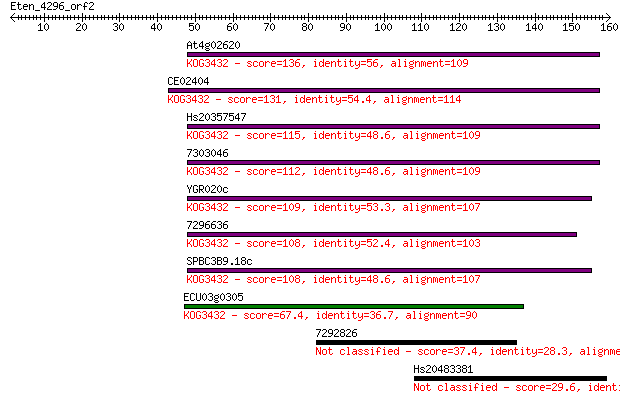
At4g02620 136 2e-32
CE02404 131 6e-31
Hs20357547 115 2e-26
7303046 112 4e-25
YGR020c 109 2e-24
7296636 108 4e-24
SPBC3B9.18c 108 5e-24
ECU03g0305 67.4 1e-11
7292826 37.4 0.014
Hs20483381 29.6 2.4
> At4g02620
Length=128
Score = 136 bits (342), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 83/109 (76%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I +I DEDTV GFLMAG+G D KTN+L+VD+KT + IE AF +F+ R D+ I+L++
Sbjct 15 IAMIADEDTVVGFLMAGVGNVDIRRKTNYLIVDSKTTVRQIEDAFKEFSARDDIAIILLS 74
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q++A+ IR+LVD +++ VP ILEIPSKD PYDP+ DSV+ RVK+ F E
Sbjct 75 QYIANMIRFLVDSYNKPVPAILEIPSKDHPYDPAHDSVLSRVKYLFSAE 123
> CE02404
Length=121
Score = 131 bits (329), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 62/115 (53%), Positives = 83/115 (72%), Gaps = 1/115 (0%)
Query 43 AAELKI-CVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDV 101
AA+ KI VIGDEDTV GFL+ G+G + K N+L+VD +T Q+IE AF+ F R D+
Sbjct 4 AAKGKILAVIGDEDTVVGFLLGGVGELNKARKPNYLIVDKQTTVQEIEEAFNGFCARDDI 63
Query 102 GIVLINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
I+LINQH+A+ IRY VD H++ +P +LEIPSK+ PYDPSKDS++ R + F E
Sbjct 64 AIILINQHIAEMIRYAVDNHTQSIPAVLEIPSKEAPYDPSKDSILNRARGLFNPE 118
> Hs20357547
Length=119
Score = 115 bits (289), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 53/109 (48%), Positives = 75/109 (68%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I VIGDEDTV GFL+ GIG + NFLVV+ T +IE F F +R D+GI+LIN
Sbjct 8 IAVIGDEDTVTGFLLGGIGELNKNRHPNFLVVEKDTTINEIEDTFRQFLNRDDIGIILIN 67
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q++A+ +R+ +D H + +P +LEIPSK+ PYD +KDS+++R + F E
Sbjct 68 QYIAEMVRHALDAHQQSIPAVLEIPSKEHPYDAAKDSILRRARGMFTAE 116
> 7303046
Length=124
Score = 112 bits (279), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 53/109 (48%), Positives = 72/109 (66%), Gaps = 0/109 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
I VIGDEDT GFL+ G+G + NF+VVD T ++E F F R D+ I+LIN
Sbjct 12 ISVIGDEDTCVGFLLGGVGEINKNRHPNFMVVDKNTAVSELEDCFKRFLKRDDIDIILIN 71
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFGGE 156
Q+ A+ IR+++D H+ VP +LEIPSKD PYD SKDS+++R + F E
Sbjct 72 QNCAELIRHVIDAHTSPVPAVLEIPSKDHPYDASKDSILRRARGMFNPE 120
> YGR020c
Length=118
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 57/110 (51%), Positives = 73/110 (66%), Gaps = 3/110 (2%)
Query 48 ICVIGDEDTVAGFLMAGIG-MRDGMGKTNFLVV-DTKTKRQDIERAFHDFTH-RSDVGIV 104
I VI DEDT G L+AGIG + + NF V + KT +++I F+ FT R D+ I+
Sbjct 8 IAVIADEDTTTGLLLAGIGQITPETQEKNFFVYQEGKTTKEEITDKFNHFTEERDDIAIL 67
Query 105 LINQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFG 154
LINQH+A+NIR VD + P ILEIPSKD PYDP KDSV++RV+ FG
Sbjct 68 LINQHIAENIRARVDSFTNAFPAILEIPSKDHPYDPEKDSVLKRVRKLFG 117
> 7296636
Length=124
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 54/103 (52%), Positives = 69/103 (66%), Gaps = 0/103 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLIN 107
+ VIGDEDT GFL+ GIG D +TNF+VV+ T + IE F F R D+ I+LIN
Sbjct 12 LAVIGDEDTCVGFLLGGIGEVDEDRETNFMVVERDTTPKQIEECFKKFLRRPDIVIILIN 71
Query 108 QHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVK 150
Q AD IR VD H+ VPT+LEIPSK PYD S+DS+++R +
Sbjct 72 QVYADMIRPTVDAHNLAVPTVLEIPSKQHPYDSSRDSILKRAQ 114
> SPBC3B9.18c
Length=120
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 52/108 (48%), Positives = 70/108 (64%), Gaps = 1/108 (0%)
Query 48 ICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFT-HRSDVGIVLI 106
+ VIGD+DTV G L+AG G + G NF ++ KT + I AF D+T R D+ IVLI
Sbjct 12 VSVIGDDDTVTGMLLAGTGQVNENGDKNFFIITQKTTDEQIAEAFDDYTTKRKDIAIVLI 71
Query 107 NQHVADNIRYLVDLHSRVVPTILEIPSKDRPYDPSKDSVMQRVKFFFG 154
NQ A+ IR ++ H + P +LEIPSKD PYDP KDS+++RV+ G
Sbjct 72 NQFAAERIRDRIENHVQAFPAVLEIPSKDDPYDPEKDSILRRVRKIIG 119
> ECU03g0305
Length=95
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 54/90 (60%), Gaps = 2/90 (2%)
Query 47 KICVIGDEDTVAGFLMAGIGMRDGMGKTNFLVVDTKTKRQDIERAFHDFTHRSDVGIVLI 106
+I +IGDE+T+ GFL+AG+ + N + V + T D+ RAF+ T R D+ IVL+
Sbjct 6 RIGIIGDEETLTGFLIAGV--ENTHDNPNLIQVASATSEDDLRRAFYSLTSREDLAIVLV 63
Query 107 NQHVADNIRYLVDLHSRVVPTILEIPSKDR 136
A+ ++ +D + +VP IL I SK++
Sbjct 64 CDFAAEKLKDEIDTYKEIVPAILVIASKNK 93
> 7292826
Length=135
Score = 37.4 bits (85), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 34/53 (64%), Gaps = 0/53 (0%)
Query 82 KTKRQDIERAFHDFTHRSDVGIVLINQHVADNIRYLVDLHSRVVPTILEIPSK 134
+T ++D+E+ F RS++GIV+I+ +R ++ S+++P ++ +P+K
Sbjct 61 ETPQEDVEQFFLTVYRRSNIGIVIIDYDTVKRLRTMMQRCSQLLPVLVTVPNK 113
> Hs20483381
Length=225
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query 108 QHVADNIRYLVDLHSRVV-PTILEIPSKDRPYDPSKDSVMQRVKFFFGGELP 158
+ +A+N+ ++L + ++ PT L +P D+ Y +D++ +R+ FG + P
Sbjct 16 REIANNLGLRIELTTPLLYPTTLALPPDDQIYQIRRDTMTKRISPLFGEQRP 67
Lambda K H
0.322 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40