bitscore colors: <40, 40-50 , 50-80, 80-200, >200
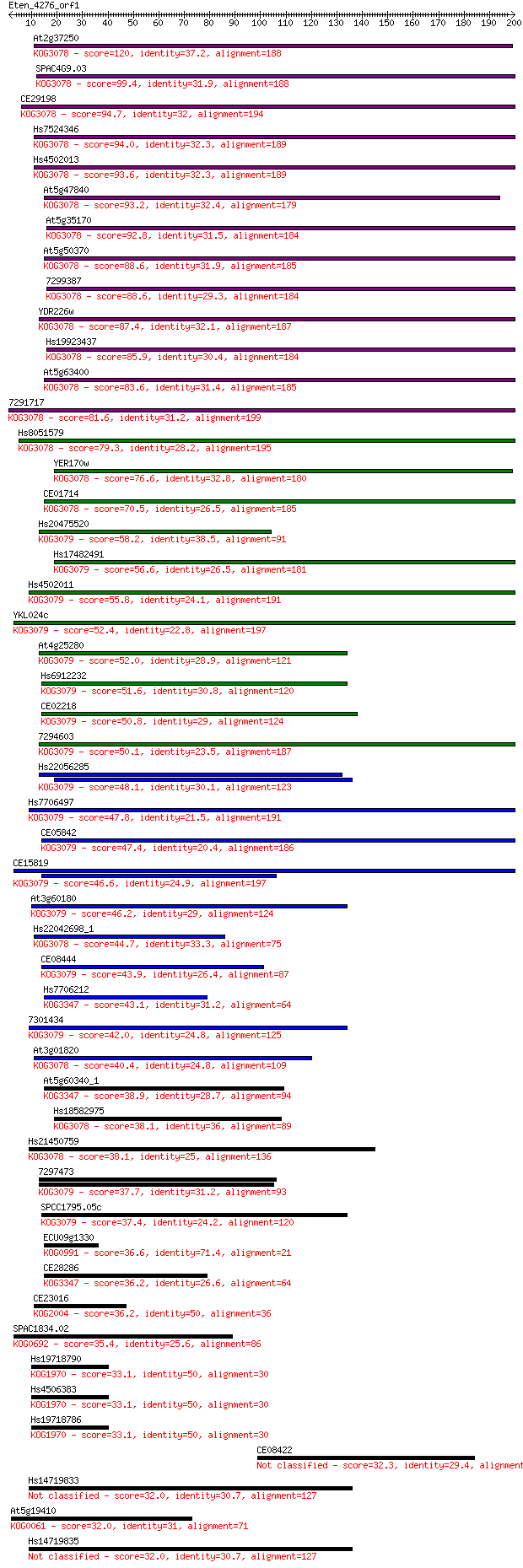
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4276_orf1
Length=199
Score E
Sequences producing significant alignments: (Bits) Value
At2g37250 120 1e-27
SPAC4G9.03 99.4 5e-21
CE29198 94.7 1e-19
Hs7524346 94.0 2e-19
Hs4502013 93.6 2e-19
At5g47840 93.2 3e-19
At5g35170 92.8 4e-19
At5g50370 88.6 8e-18
7299387 88.6 8e-18
YDR226w 87.4 2e-17
Hs19923437 85.9 5e-17
At5g63400 83.6 3e-16
7291717 81.6 8e-16
Hs8051579 79.3 5e-15
YER170w 76.6 3e-14
CE01714 70.5 2e-12
Hs20475520 58.2 1e-08
Hs17482491 56.6 3e-08
Hs4502011 55.8 5e-08
YKL024c 52.4 5e-07
At4g25280 52.0 7e-07
Hs6912232 51.6 1e-06
CE02218 50.8 2e-06
7294603 50.1 3e-06
Hs22056285 48.1 1e-05
Hs7706497 47.8 1e-05
CE05842 47.4 2e-05
CE15819 46.6 4e-05
At3g60180 46.2 4e-05
Hs22042698_1 44.7 1e-04
CE08444 43.9 2e-04
Hs7706212 43.1 3e-04
7301434 42.0 7e-04
At3g01820 40.4 0.003
At5g60340_1 38.9 0.008
Hs18582975 38.1 0.012
Hs21450759 38.1 0.013
7297473 37.7 0.015
SPCC1795.05c 37.4 0.020
ECU09g1330 36.6 0.037
CE28286 36.2 0.041
CE23016 36.2 0.042
SPAC1834.02 35.4 0.085
Hs19718790 33.1 0.38
Hs4506383 33.1 0.40
Hs19718786 33.1 0.42
CE08422 32.3 0.58
Hs14719833 32.0 0.81
At5g19410 32.0 0.83
Hs14719835 32.0 0.87
> At2g37250
Length=284
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/197 (35%), Positives = 109/197 (55%), Gaps = 14/197 (7%)
Query 11 KGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARVSCGLL 69
+ + +F G PG GKGTY L+ G+ H++ GDL+R++L S+ LS V+ G L
Sbjct 50 RNVQWVFLGCPGVGKGTYASRLSTLLGVPHIATGDLVREELASSGPLSQKLSEIVNQGKL 109
Query 70 IQDTLASEICKDYISSHPER--QSWILDGFPRTLQQVELLRGFATPHACINFIVPRHILL 127
+ D + ++ + + R +ILDGFPRT++Q E+L +N +P +L+
Sbjct 110 VSDEIIVDLLSKRLEAGEARGESGFILDGFPRTMRQAEILGDVTDIDLVVNLKLPEEVLV 169
Query 128 KKLLGRRICSSCGGCFNVANICDEPYD------MPPILPTKECRHCKGNAPLTTRSDDTE 181
K LGRR CS CG FNVA+I + + M P+LP +C + L TR+DDTE
Sbjct 170 DKCLGRRTCSQCGKGFNVAHINLKGENGRPGISMDPLLPPHQCM-----SKLVTRADDTE 224
Query 182 EAIRNRLDFYEQLTRPM 198
E ++ RL Y + ++P+
Sbjct 225 EVVKARLRIYNETSQPL 241
> SPAC4G9.03
Length=220
Score = 99.4 bits (246), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 60/195 (30%), Positives = 105/195 (53%), Gaps = 17/195 (8%)
Query 12 GIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSNDSEARVSCGLLI 70
G+ LI GPPG+GKGT + +++G+ H++ GD++R +++ T L +++ + G L+
Sbjct 3 GMRLILVGPPGAGKGTQAPNIQKKYGIAHLATGDMLRSQVARQTELGKEAKKIMDQGGLV 62
Query 71 QDTLASEICKDYISSHPE-RQSWILDGFPRTLQQVELLRGFATP-----HACINFIVPRH 124
D + + + KD I ++PE + +ILDGFPRT+ Q E L + + V
Sbjct 63 SDDIVTGMIKDEILNNPECKNGFILDGFPRTVVQAEKLTALLDELKLDLNTVLELQVDDE 122
Query 125 ILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAI 184
+L++++ GR + G +++ PP +P K+ G PL RSDD +A+
Sbjct 123 LLVRRITGRLVHPGSGRSYHLEF-------NPPKVPMKD--DVTGE-PLIQRSDDNADAL 172
Query 185 RNRLDFYEQLTRPML 199
R RL Y + T P++
Sbjct 173 RKRLVTYHEQTTPVV 187
> CE29198
Length=251
Score = 94.7 bits (234), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 62/200 (31%), Positives = 102/200 (51%), Gaps = 16/200 (8%)
Query 6 STCVQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARV 64
+ + +GI IF GPPGSGKGT A+++ H++ GDL+R ++ S + +A +
Sbjct 20 TETLARGIRAIFIGPPGSGKGTQAPAFAQKYFSCHLATGDLLRAEVASGSEFGKELKATM 79
Query 65 SCGLLIQDTLASEICKDYISSHPERQSWILDGFPRTLQQV----ELLRGFATP-HACINF 119
G L+ D + ++ + + + +ILDGFPRT Q E+L TP + F
Sbjct 80 DAGKLVSDEVVCKLIEQKLEKPECKYGFILDGFPRTSGQAEKLDEILERRKTPLDTVVEF 139
Query 120 IVPRHILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDD 179
+ +L++++ GR + G +++ PP +P K+ G PL RSDD
Sbjct 140 NIADDLLVRRITGRLFHIASGRSYHLEF-------KPPKVPMKD--DLTGE-PLIRRSDD 189
Query 180 TEEAIRNRLDFYEQLTRPML 199
EE +R RL Y Q+T P++
Sbjct 190 NEETLRKRLVQYHQMTVPLV 209
> Hs7524346
Length=232
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 61/195 (31%), Positives = 101/195 (51%), Gaps = 16/195 (8%)
Query 11 KGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARVSCGLL 69
KGI + GPPG+GKGT LA + + H++ GD++R + S L +A + G L
Sbjct 14 KGIRAVLLGPPGAGKGTQAPRLAENFCVCHLATGDMLRAMVASGSELGKKLKATMDAGKL 73
Query 70 IQDTLASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPH-----ACINFIVPRH 124
+ D + E+ + + + + ++LDGFPRT++Q E+L + I F +P
Sbjct 74 VSDEMVVELIEKNLETPLCKNGFLLDGFPRTVRQAEMLDDLMEKRKEKLDSVIEFSIPDS 133
Query 125 ILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAI 184
+L++++ GR I G ++ E ++ PP P K+ G PL RSDD E+A+
Sbjct 134 LLIRRITGRLIHPKSGRSYH------EEFN-PPKEPMKD--DITGE-PLIRRSDDNEKAL 183
Query 185 RNRLDFYEQLTRPML 199
+ RL Y T P++
Sbjct 184 KIRLQAYHTQTTPLI 198
> Hs4502013
Length=239
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 61/195 (31%), Positives = 101/195 (51%), Gaps = 16/195 (8%)
Query 11 KGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARVSCGLL 69
KGI + GPPG+GKGT LA + + H++ GD++R + S L +A + G L
Sbjct 14 KGIRAVLLGPPGAGKGTQAPRLAENFCVCHLATGDMLRAMVASGSELGKKLKATMDAGKL 73
Query 70 IQDTLASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPH-----ACINFIVPRH 124
+ D + E+ + + + + ++LDGFPRT++Q E+L + I F +P
Sbjct 74 VSDEMVVELIEKNLETPLCKNGFLLDGFPRTVRQAEMLDDLMEKRKEKLDSVIEFSIPDS 133
Query 125 ILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAI 184
+L++++ GR I G ++ E ++ PP P K+ G PL RSDD E+A+
Sbjct 134 LLIRRITGRLIHPKSGRSYH------EEFN-PPKEPMKD--DITGE-PLIRRSDDNEKAL 183
Query 185 RNRLDFYEQLTRPML 199
+ RL Y T P++
Sbjct 184 KIRLQAYHTQTTPLI 198
> At5g47840
Length=283
Score = 93.2 bits (230), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 58/183 (31%), Positives = 97/183 (53%), Gaps = 19/183 (10%)
Query 15 LIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEAR--VSCGLLIQD 72
++ SG P SGKGT +++ ++G+ H+S GDL+R ++++ + N A+ + G L+ D
Sbjct 67 IMISGAPASGKGTQCELITHKYGLVHISAGDLLRAEIASGS-ENGRRAKEHMEKGQLVPD 125
Query 73 TLASEICKDYIS-SHPERQSWILDGFPRTLQQVELLRGFA-TPHACINFIVPRHILLKKL 130
+ + KD +S + E++ W+LDG+PR+ Q L+GF P I VP IL++++
Sbjct 126 EIVVMMVKDRLSQTDSEQKGWLLDGYPRSASQATALKGFGFQPDLFIVLEVPEEILIERV 185
Query 131 LGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRLDF 190
+GRR+ G +++ PP R LT R DDTEE + RL
Sbjct 186 VGRRLDPVTGKIYHLKY-------SPPETEEIAVR-------LTQRFDDTEEKAKLRLKT 231
Query 191 YEQ 193
+ Q
Sbjct 232 HNQ 234
> At5g35170
Length=498
Score = 92.8 bits (229), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 58/187 (31%), Positives = 97/187 (51%), Gaps = 17/187 (9%)
Query 16 IFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSNDSEARVSCGLLIQDTL 74
+ SG P SGKGT +++ ++G+ H+S GDL+R ++S+ T + ++ ++ G L+ D +
Sbjct 1 MISGAPASGKGTQCELIVHKFGLVHISTGDLLRAEVSSGTDIGKRAKEFMNSGSLVPDEI 60
Query 75 ASEICKDYISSHPERQ-SWILDGFPRTLQQVELLRGF-ATPHACINFIVPRHILLKKLLG 132
+ +S ++ W+LDGFPR+ Q + L P I VP IL+ + +G
Sbjct 61 VIAMVAGRLSREDAKEHGWLLDGFPRSFAQAQSLDKLNVKPDIFILLDVPDEILIDRCVG 120
Query 133 RRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRLDFYE 192
RR+ G +++ N PP E K A L TR DDTEE ++ RL Y+
Sbjct 121 RRLDPVTGKIYHIKN-------YPP-----ESDEIK--ARLVTRPDDTEEKVKARLQIYK 166
Query 193 QLTRPML 199
Q + ++
Sbjct 167 QNSEAII 173
> At5g50370
Length=248
Score = 88.6 bits (218), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 59/191 (30%), Positives = 97/191 (50%), Gaps = 16/191 (8%)
Query 15 LIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSNDSEARVSCGLLIQDT 73
L+F GPPGSGKGT ++ + + H+S GD++R ++ T L ++ + G L+ D
Sbjct 37 LVFIGPPGSGKGTQSPVIKDEFCLCHLSTGDMLRAAVAAKTPLGVKAKEAMDKGELVSDD 96
Query 74 LASEICKDYISSHPERQSWILDGFPRTLQQVELL-----RGFATPHACINFIVPRHILLK 128
L I + ++ ++ +ILDGFPRT+ Q E L R A +NF + +L +
Sbjct 97 LVVGIMDEAMNRPKCQKGFILDGFPRTVTQAEKLDEMLNRRGAQIDKVLNFAIDDSVLEE 156
Query 129 KLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRL 188
++ GR I S G ++ PP +P G PL R DD + +R+RL
Sbjct 157 RITGRWIHPSSGRSYHTKF-------APPKVPG--VDDLTGE-PLIQRKDDNADVLRSRL 206
Query 189 DFYEQLTRPML 199
D + + T+P++
Sbjct 207 DAFHKQTQPVI 217
> 7299387
Length=216
Score = 88.6 bits (218), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 54/185 (29%), Positives = 91/185 (49%), Gaps = 12/185 (6%)
Query 16 IFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQK-LSNCTLSNDSEARVSCGLLIQDTL 74
+ G PGSGKGT +++ + G H+S GD++RQ + N L ++ ++ G L+ D +
Sbjct 9 VIIGAPGSGKGTISELICKNHGCVHISTGDILRQNIIKNTELGKKAKQYIAEGKLVPDAI 68
Query 75 ASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPHACINFIVPRHILLKKLLGRR 134
++ I+ R S+ILDGFPR + Q E L A I VP +++ ++ R
Sbjct 69 VTKTMLARITEVGNR-SYILDGFPRNIAQAEALAAREQIDAVITLDVPHSVIIDRVKNRW 127
Query 135 ICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRLDFYEQL 194
I + G +N+ P +P K+ G PL R DD + RL+ Y+++
Sbjct 128 IHAPSGRVYNIGF-------KNPKVPGKD--DVTGE-PLMQREDDKPHVVAKRLELYDEV 177
Query 195 TRPML 199
P++
Sbjct 178 MSPVI 182
> YDR226w
Length=222
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 60/195 (30%), Positives = 101/195 (51%), Gaps = 19/195 (9%)
Query 13 IILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSNDSEARVSCGLLIQ 71
I ++ GPPG+GKGT L R+ H++ GD++R +++ T L +++ + G L+
Sbjct 7 IRMVLIGPPGAGKGTQAPNLQERFHAAHLATGDMLRSQIAKGTQLGLEAKKIMDQGGLVS 66
Query 72 DTLASEICKDYISSHPE-RQSWILDGFPRTLQQVE----LLRGFATP-HACINFIVPRHI 125
D + + KD ++++P + +ILDGFPRT+ Q E +L+ TP I V +
Sbjct 67 DDIMVNMIKDELTNNPACKNGFILDGFPRTIPQAEKLDQMLKEQGTPLEKAIELKVDDEL 126
Query 126 LLKKLLGRRICSSCGGCFN-VANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAI 184
L+ ++ GR I + G ++ + N PP K+ G A L RSDD +A+
Sbjct 127 LVARITGRLIHPASGRSYHKIFN--------PPKEDMKD--DVTGEA-LVQRSDDNADAL 175
Query 185 RNRLDFYEQLTRPML 199
+ RL Y T P++
Sbjct 176 KKRLAAYHAQTEPIV 190
> Hs19923437
Length=227
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 56/185 (30%), Positives = 89/185 (48%), Gaps = 12/185 (6%)
Query 16 IFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSNDSEARVSCGLLIQDTL 74
+ G PGSGKGT + + ++H+S GDL+R + T + ++A + G LI D +
Sbjct 11 VIMGAPGSGKGTVSSRITTHFELKHLSSGDLLRDNMLRGTEIGVLAKAFIDQGKLIPDDV 70
Query 75 ASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPHACINFIVPRHILLKKLLGRR 134
+ + + + + SW+LDGFPRTL Q E L IN VP ++ ++L R
Sbjct 71 MTRLALHELKNLTQ-YSWLLDGFPRTLPQAEALDRAYQIDTVINLNVPFEVIKQRLTARW 129
Query 135 ICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRLDFYEQL 194
I + G +N+ ++ P + + PL R DD E + RL YE
Sbjct 130 IHPASGRVYNIE------FNPPKTVGIDDLT----GEPLIQREDDKPETVIKRLKAYEDQ 179
Query 195 TRPML 199
T+P+L
Sbjct 180 TKPVL 184
> At5g63400
Length=246
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 58/191 (30%), Positives = 94/191 (49%), Gaps = 16/191 (8%)
Query 15 LIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSNDSEARVSCGLLIQDT 73
LIF GPPGSGKGT ++ + + H+S GD++R +++ T L ++ + G L+ D
Sbjct 36 LIFIGPPGSGKGTQSPVVKDEYCLCHLSTGDMLRAAVASKTPLGVKAKEAMEKGELVSDD 95
Query 74 LASEICKDYISSHPERQSWILDGFPRTLQQVE-----LLRGFATPHACINFIVPRHILLK 128
L I + ++ ++ +ILDGFPRT+ Q E L R +NF + IL +
Sbjct 96 LVVGIIDEAMNKPKCQKGFILDGFPRTVTQAEKLDEMLKRRGTEIDKVLNFAIDDAILEE 155
Query 129 KLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRL 188
++ GR I S G ++ PP P G PL R DD + +++RL
Sbjct 156 RITGRWIHPSSGRSYHTKF-------APPKTPG--VDDITGE-PLIQRKDDNADVLKSRL 205
Query 189 DFYEQLTRPML 199
+ T+P++
Sbjct 206 AAFHSQTQPVI 216
> 7291717
Length=237
Score = 81.6 bits (200), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 62/209 (29%), Positives = 104/209 (49%), Gaps = 31/209 (14%)
Query 1 RHEPLSTCVQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSND 59
R+EP + GI I GPPGSGKGT ++ + H+S GD++R ++S+ + L +
Sbjct 11 RYEPENI----GINAILLGPPGSGKGT-------QFCVCHLSTGDMLRAEISSGSKLGAE 59
Query 60 SEARVSCGLLIQDTLASEICKDYISSHPERQSWILDGFPRTLQQVE---------LLRGF 110
+ + G L+ D L ++ + + ++LDGFPRT+ Q E L +
Sbjct 60 LKKVMDAGKLVSDDLVVDMIDSNLDKPECKNGFLLDGFPRTVVQAEKVATRLDTLLDKRK 119
Query 111 ATPHACINFIVPRHILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGN 170
A I F + +L++++ GR I + G ++ E + PP P + G
Sbjct 120 TNLDAVIEFAIDDSLLVRRITGRLIHQASGRSYH------EEF-APPKKPMTD--DVTGE 170
Query 171 APLTTRSDDTEEAIRNRLDFYEQLTRPML 199
PL RSDD EA++ RL+ Y + T+P++
Sbjct 171 -PLIRRSDDNAEALKKRLEAYHKQTKPLV 198
> Hs8051579
Length=223
Score = 79.3 bits (194), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 55/196 (28%), Positives = 94/196 (47%), Gaps = 15/196 (7%)
Query 5 LSTCVQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCT-LSNDSEAR 63
+++ + + +IL GPPGSGKGT + +A+ +G++H+S G +R+ + T + ++
Sbjct 1 MASKLLRAVIL---GPPGSGKGTVCQRIAQNFGLQHLSSGHFLRENIKASTEVGEMAKQY 57
Query 64 VSCGLLIQDTLASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPHACINFIVPR 123
+ LL+ D + + + + + Q W+LDGFPRTL Q E L I+ +P
Sbjct 58 IEKSLLVPDHVITRLMMSELENR-RGQHWLLDGFPRTLGQAEALDKICEVDLVISLNIPF 116
Query 124 HILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEA 183
L +L R I G +N+ D P P PL + DD EA
Sbjct 117 ETLKDRLSRRWIHPPSGRVYNL--------DFNP--PHVHGIDDVTGEPLVQQEDDKPEA 166
Query 184 IRNRLDFYEQLTRPML 199
+ RL Y+ + +P++
Sbjct 167 VAARLRQYKDVAKPVI 182
> YER170w
Length=225
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 59/191 (30%), Positives = 94/191 (49%), Gaps = 22/191 (11%)
Query 19 GPPGSGKGTY-GKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARVSCGLLIQDTLAS 76
G PGSGKGT ++L + + +S GD++RQ++ S TL ++ ++ G L+ D L +
Sbjct 21 GAPGSGKGTQTSRLLKQIPQLSSISSGDILRQEIKSESTLGREATTYIAQGKLLPDDLIT 80
Query 77 EICKDYISS----HPERQSWILDGFPRTLQQVELLRGFATPH-ACINFIV----PRHILL 127
+ +S+ P W+LDGFPRT Q L H A +N +V P +L
Sbjct 81 RLITFRLSALGWLKPSAM-WLLDGFPRTTAQASALDELLKQHDASLNLVVELDVPESTIL 139
Query 128 KKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNR 187
+++ R + G +N+ Y+ PP +P G PLT R DDT E + R
Sbjct 140 ERIENRYVHVPSGRVYNLQ------YN-PPKVPG--LDDITGE-PLTKRLDDTAEVFKKR 189
Query 188 LDFYEQLTRPM 198
L+ Y++ P+
Sbjct 190 LEEYKKTNEPL 200
> CE01714
Length=222
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/190 (25%), Positives = 93/190 (48%), Gaps = 18/190 (9%)
Query 15 LIFSGPPGSGKGTYGKILARRW---GMRHVSLGDLIRQKLSNCT-LSNDSEARVSCGLLI 70
++ SG GSGKGT ++L R + G + + GD IR ++ T +++ ++ G +
Sbjct 4 VLLSGAAGSGKGTIARMLVREFEPLGFNYFAAGDFIRDHIARGTEFGVRAQSFLNKGEHV 63
Query 71 QDT-LASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPHACINFIVPRHILLKK 129
D+ L I + + + P +LDG+PR + Q++++ A + + VPR +L+ +
Sbjct 64 PDSILNGAILAEMLKAGPR---VVLDGYPRNMSQLKMVEEQAPLNLIVELKVPRKVLIDR 120
Query 130 LLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRLD 189
L + + + G +N+ ++ P P +E + PL RS D E R RL+
Sbjct 121 LSKQLVHPASGRAYNL--------EVNP--PKEEGKDDITGEPLFKRSTDQLEVARRRLE 170
Query 190 FYEQLTRPML 199
Y++ +L
Sbjct 171 VYDKTENKVL 180
> Hs20475520
Length=166
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/95 (36%), Positives = 57/95 (60%), Gaps = 6/95 (6%)
Query 13 IILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL----SNCTLSNDSEARVSCGL 68
IIL+ GP GSGKGT +A R+G +++S+G+L+R+K+ SN S ++ + L
Sbjct 53 IILVIGGP-GSGKGTQSLKIAERYGFQYISVGELLRKKIHSTSSNRKWSLIAKIITTGEL 111
Query 69 LIQDTLASEICKDYISSHPERQSWILDGFPRTLQQ 103
Q+T +EI K + P+ + ++DGFPR + Q
Sbjct 112 APQETTITEI-KQKLMQIPDEEGIVIDGFPRDVAQ 145
> Hs17482491
Length=165
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 48/183 (26%), Positives = 81/183 (44%), Gaps = 29/183 (15%)
Query 19 GPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEARVSC-GLLIQDTLASE 77
G PG GKGT K +A ++G HV L L+RQ+ T ++ GLL+ + +
Sbjct 2 GGPGCGKGTQCKNMATKYGFCHVGLDQLLRQEAQRSTQRGRQIRDITLQGLLVPAGIIPD 61
Query 78 ICKDYISSHPERQSWILDGFPRTLQQ-VELLRGFATPHACINFIVPRHILLKKLLGRRIC 136
+ D + S PE + +++DGFP+ ++Q +E R + P + +++
Sbjct 62 MVSDNMLSRPESRGFLIDGFPQEVKQAMEFERIVSGPEVWVWVGQAPSVVI--------- 112
Query 137 SSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRLDFYEQLTR 196
F+ C + +L + H R+DD+E AI RLD + L
Sbjct 113 -----VFD----CSMETMLRRVLHWGQVEH---------RADDSELAIHQRLDTHYTLCE 154
Query 197 PML 199
P+L
Sbjct 155 PVL 157
> Hs4502011
Length=194
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 46/194 (23%), Positives = 81/194 (41%), Gaps = 40/194 (20%)
Query 9 VQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSND--SEARVSC 66
++K I+ G PGSGKGT + + +++G H+S GDL+R ++S+ + SE
Sbjct 5 LKKTKIIFVVGGPGSGKGTQCEKIVQKYGYTHLSTGDLLRSEVSSGSARGKKLSEIMEKG 64
Query 67 GLLIQDTLASEICKDYISSHPERQSWILDGFPRTLQQ-VELLRGFATPHACINFIVPRHI 125
L+ +T+ + ++ + +++DG+PR +QQ E R P +
Sbjct 65 QLVPLETVLDMLRDAMVAKVNTSKGFLIDGYPREVQQGEEFERRIGQPTLLLYVDAGPET 124
Query 126 LLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIR 185
+ ++LL R S R DD EE I+
Sbjct 125 MTQRLLKRGETSG-------------------------------------RVDDNEETIK 147
Query 186 NRLDFYEQLTRPML 199
RL+ Y + T P++
Sbjct 148 KRLETYYKATEPVI 161
> YKL024c
Length=204
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 45/203 (22%), Positives = 84/203 (41%), Gaps = 45/203 (22%)
Query 3 EPLSTCVQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEA 62
+P + Q +I + GP G+GKGT + L + + H+S GDL+R + S E
Sbjct 8 QPAFSPDQVSVIFVLGGP-GAGKGTQCEKLVKDYSFVHLSAGDLLRAEQGRAG-SQYGEL 65
Query 63 RVSC---GLLIQDTLASEICKDYISSH--PERQSWILDGFPRTLQQ-VELLRGFATPHAC 116
+C G ++ + + ++ IS + + +++DGFPR + Q + R
Sbjct 66 IKNCIKEGQIVPQEITLALLRNAISDNVKANKHKFLIDGFPRKMDQAISFERDIVESKFI 125
Query 117 INFIVPRHILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTR 176
+ F P I+L++LL R + R
Sbjct 126 LFFDCPEDIMLERLLER-------------------------------------GKTSGR 148
Query 177 SDDTEEAIRNRLDFYEQLTRPML 199
SDD E+I+ R + +++ + P++
Sbjct 149 SDDNIESIKKRFNTFKETSMPVI 171
> At4g25280
Length=227
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 66/124 (53%), Gaps = 4/124 (3%)
Query 13 IILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEAR-VSCGLLIQ 71
I G PGSGKGT + + +G++H+S GDL+R++++ T + + G ++
Sbjct 44 FITFVLGGPGSGKGTQCEKIVETFGLQHLSAGDLLRREIAMHTENGAMILNLIKDGKIVP 103
Query 72 DTLASEICKDYISSHPERQSWILDGFPRTLQ-QVELLRGF-ATPHACINFIVPRHILLKK 129
+ ++ + + S R+ +++DGFPRT + +V R A P + F P ++K+
Sbjct 104 SEVTVKLIQKELESSDNRK-FLIDGFPRTEENRVAFERIIRADPDVVLFFDCPEEEMVKR 162
Query 130 LLGR 133
+L R
Sbjct 163 VLNR 166
> Hs6912232
Length=198
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 64/126 (50%), Gaps = 9/126 (7%)
Query 14 ILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSN----CTLSNDSEARVSCGLL 69
I+ G PGSGKGT + L ++G H+S G+L+R++L++ L D R G L
Sbjct 13 IIFIIGGPGSGKGTQCEKLVEKYGFTHLSTGELLREELASESERSKLIRDIMER---GDL 69
Query 70 IQDTLASEICKD-YISSHPERQSWILDGFPRTLQQVELL-RGFATPHACINFIVPRHILL 127
+ + E+ K+ ++S + + +++DG+PR ++Q E R P I +
Sbjct 70 VPSGIVLELLKEAMVASLGDTRGFLIDGYPREVKQGEEFGRRIGDPQLVICMDCSADTMT 129
Query 128 KKLLGR 133
+LL R
Sbjct 130 NRLLQR 135
> CE02218
Length=210
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 61/128 (47%), Gaps = 4/128 (3%)
Query 14 ILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARVSCGLLIQD 72
I G PGSGKGT + ++G+ H+S GDL+R ++ S A + G L+
Sbjct 22 IFFIVGGPGSGKGTQCDKIVAKYGLTHLSSGDLLRDEVKSGSPRGAQLTAIMESGALVPL 81
Query 73 TLASEICKDYISSHPERQS--WILDGFPRTLQQVELLRG-FATPHACINFIVPRHILLKK 129
+ ++ K+ + E+ S +++DG+PR + Q + + F V L+K+
Sbjct 82 EVVLDLVKEAMLKAIEKGSKGFLIDGYPREVAQGQQFESEIQEAKLVLFFDVAEETLVKR 141
Query 130 LLGRRICS 137
LL R S
Sbjct 142 LLHRAQTS 149
> 7294603
Length=201
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 44/190 (23%), Positives = 77/190 (40%), Gaps = 39/190 (20%)
Query 13 IILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARVSCGLLIQ 71
II I GP G GKGT + ++G H+S GDL+R ++ S +A ++ G L+
Sbjct 6 IIWILGGP-GCGKGTQCAKIVEKYGFTHLSSGDLLRNEVASGSDKGRQLQAVMASGGLVS 64
Query 72 DTLASEICKDYIS-SHPERQSWILDGFPRTLQQ-VELLRGFATPHACINFIVPRHILLKK 129
+ + D I+ + + +++DG+PR Q +E A + F ++++
Sbjct 65 NDEVLSLLNDAITRAKGSSKGFLIDGYPRQKNQGIEFEARIAPADLALYFECSEDTMVQR 124
Query 130 LLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNRLD 189
++ R S+ R DD E+ IR RL
Sbjct 125 IMARAAASA-----------------------------------VKRDDDNEKTIRARLL 149
Query 190 FYEQLTRPML 199
++Q T +L
Sbjct 150 TFKQNTNAIL 159
> Hs22056285
Length=826
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 36/124 (29%), Positives = 58/124 (46%), Gaps = 5/124 (4%)
Query 13 IILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSN---CTLSNDSEARVSCGLL 69
I +I GPP SGK T K + +G++H+S+G +R L+N L+ + G+
Sbjct 491 IRIIIVGPPKSGKTTVAKKITSEYGLKHLSIGGALRYVLNNHPETELALMLNWHLHKGMT 550
Query 70 IQDTLASEICK-DYISSHPERQSWILDGFPRTLQQVELLRGFA-TPHACINFIVPRHILL 127
D LA + + + S ++DG+P T Q+ LL + P VP +
Sbjct 551 APDELAIQALELSLMESVCNTAGVVIDGYPVTKHQMNLLEARSIIPMVIFELSVPSKEIF 610
Query 128 KKLL 131
K+LL
Sbjct 611 KRLL 614
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 38/169 (22%), Positives = 65/169 (38%), Gaps = 53/169 (31%)
Query 19 GPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL--------------------------- 51
GP GSGK G+ LA + + H+ +++++KL
Sbjct 77 GPQGSGKTMCGRQLAEKLNIFHIQFEEVLQEKLLLKTEKKVGPEFEEDSENEQAAKQELE 136
Query 52 -----SNCTLSNDS-------------EARVSCGLLIQDTLASEICKDYISS----HPER 89
+N + ++ E + L+ + L EI + +S P R
Sbjct 137 ELAIQANVKVEEENTKKQLPEVQLTEEEEVIKSSLMENEPLPPEILEVILSEWWLKEPIR 196
Query 90 QS-WILDGFPRTLQQVELL--RGFATPHACINFIVPRHILLKKLLGRRI 135
+ +ILDGFPR ++ + L RGF P A + V + +LL +I
Sbjct 197 STGFILDGFPRYPEEAQFLGDRGF-FPDAAVFIQVDDQDIFDRLLPAQI 244
> Hs7706497
Length=228
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/198 (20%), Positives = 81/198 (40%), Gaps = 41/198 (20%)
Query 9 VQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSN--CTLSNDSEARVSC 66
+ K +++ G PG+GKGT + ++G H+S G+L+R + N E +
Sbjct 32 LMKPLVVFVLGGPGAGKGTQCARIVEKYGYTHLSAGELLRDERKNPDSQYGELIEKYIKE 91
Query 67 GLLIQDTLASEICK----DYISSHPERQSWILDGFPRTLQQVELLRGFATPHACINFIVP 122
G ++ + + K ++++ ++ +++DGFPR ++ A ++F++
Sbjct 92 GKIVPVEITISLLKREMDQTMAANAQKNKFLIDGFPRNQDNLQGWNKTMDGKADVSFVL- 150
Query 123 RHILLKKLLGRRICSSCGGCFNVAN-ICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTE 181
F+ N IC E C + RSDD
Sbjct 151 -------------------FFDCNNEICIE--------------RCLERGKSSGRSDDNR 177
Query 182 EAIRNRLDFYEQLTRPML 199
E++ R+ Y Q T+P++
Sbjct 178 ESLEKRIQTYLQSTKPII 195
> CE05842
Length=191
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 38/192 (19%), Positives = 76/192 (39%), Gaps = 45/192 (23%)
Query 14 ILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIR--QKLSNCTLSNDSEARVSCGLLIQ 71
++ GPPGSGKGT + H+S GDL+R ++ E+ + G ++
Sbjct 4 VVFVLGPPGSGKGTICAKIQENLNYVHLSAGDLLRAERQREGSEFGALIESHIKNGSIVP 63
Query 72 DTLASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPHACINFIV----PRHILL 127
+ + ++ + + + + +++DGFPR ++ A + F++ P I +
Sbjct 64 VEITCSLLENAMKACGDAKGFLVDGFPRNEDNLQGWNKQMDGKALVQFVLFLSCPVSICI 123
Query 128 KKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTRSDDTEEAIRNR 187
++ L R R+DD EE+++ R
Sbjct 124 ERCLNRG---------------------------------------QGRTDDNEESLKKR 144
Query 188 LDFYEQLTRPML 199
++ Y Q T P++
Sbjct 145 VETYNQQTFPII 156
> CE15819
Length=736
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 49/203 (24%), Positives = 93/203 (45%), Gaps = 44/203 (21%)
Query 3 EPLSTCVQKGIILIFSGPPGSGKGTYGKILARRW-GMRHVSLGDLIRQKLSNCTLSNDSE 61
EP+ +IL+ G PGS K + +A+++ G +S+GD++R+K++N +++
Sbjct 505 EPVGLPNNAPVILVL-GAPGSQKNDISRRIAQKYDGFTMLSMGDILRKKINN-EKNDEMW 562
Query 62 ARVSCGLLIQDTLASEICKD--YISSHPERQS---WILDGFPRTLQQVELLRGFATPHAC 116
+VS + D + +++C+ Y H S ++++G+P++ Q+ L
Sbjct 563 DKVSKKMNNGDPIPTKMCRTVLYEELHSRGTSNWGYVIEGYPKSPDQLVDL--------- 613
Query 117 INFIVPRHILLKKLLGRRICSSCGGCFNVANICDEPYDMPPILPTKECRHCKGNAPLTTR 176
H L + L I + C V N K R K R
Sbjct 614 ------EHSLQRTDLAILIDCTEQFCLEVIN--------------KRNRENK-------R 646
Query 177 SDDTEEAIRNRLDFYEQLTRPML 199
SDD +A+R+R++++++ T PML
Sbjct 647 SDDDSDAVRSRMEYFKKNTLPML 669
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 49/102 (48%), Gaps = 13/102 (12%)
Query 14 ILIFSGPPGSGKGTYGKILARRW---GMRHVSLGDLIR-------QKLSNCTLSNDSEAR 63
I++F G PG GK + +A G+ H+ + D+IR K +N+ R
Sbjct 171 IILFMGGPGGGKTRHAARVADSLADNGLVHICMPDIIRTALGKYKDKYPEWKEANEHYIR 230
Query 64 VSCGLLIQDTLASEICKDYISSHPERQSWILDGFPRTLQQVE 105
G LI + LA + K + HP+ + L+G+PR +QVE
Sbjct 231 ---GELIPNQLALTLLKAEMGRHPDAMGFFLEGYPREARQVE 269
> At3g60180
Length=210
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 61/137 (44%), Gaps = 24/137 (17%)
Query 10 QKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEARVSCGLL 69
+K ++ G PGSGKGT + + + H S GDL+R ++ + + G +
Sbjct 25 KKSTVVFVLGGPGSGKGTQCANVVKHFSYTHFSAGDLLRAEIKSGS---------EFGAM 75
Query 70 IQDTLA------SEI-----CKDYISSHPERQSWILDGFPRTLQQVELLRGFA--TPHAC 116
IQ +A SEI CK S ++ +++DGFPR + + A P
Sbjct 76 IQSMIAEGRIVPSEITVKLLCKAMEESGNDK--FLIDGFPRNEENRNVFENVARIEPAFV 133
Query 117 INFIVPRHILLKKLLGR 133
+ F P L ++++ R
Sbjct 134 LFFDCPEEELERRIMSR 150
> Hs22042698_1
Length=110
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 38/76 (50%), Gaps = 1/76 (1%)
Query 11 KGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKL-SNCTLSNDSEARVSCGLL 69
KGI + GPPG GKGT ILA + + H + GD +R + S L A + G L
Sbjct 14 KGIQAVLLGPPGDGKGTQAPILAENFCVCHSATGDRLRAMVASGSELEKKLNATMDAGKL 73
Query 70 IQDTLASEICKDYISS 85
+ D + E+ + + +
Sbjct 74 VSDEMVVELIEKNLET 89
> CE08444
Length=191
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 43/89 (48%), Gaps = 2/89 (2%)
Query 14 ILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDS--EARVSCGLLIQ 71
++ GPPGSGKGT + G H+S GDL+R + + E + G ++
Sbjct 4 VVFVLGPPGSGKGTICTQIHENLGYVHLSAGDLLRAERERAGSEYGALIEGHIKNGSIVP 63
Query 72 DTLASEICKDYISSHPERQSWILDGFPRT 100
+ + ++ + + + +++DGFPR
Sbjct 64 VEITCALLENAMIASKDANGFLIDGFPRN 92
> Hs7706212
Length=172
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 38/64 (59%), Gaps = 4/64 (6%)
Query 15 LIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEARVSCGLLIQDTL 74
++ +G PG GK T GK LA + G++++++GDL R++ L + + C +L +D +
Sbjct 6 ILLTGTPGVGKTTLGKELASKSGLKYINVGDLAREE----QLYDGYDEEYDCPILDEDRV 61
Query 75 ASEI 78
E+
Sbjct 62 VDEL 65
> 7301434
Length=253
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 31/131 (23%), Positives = 61/131 (46%), Gaps = 7/131 (5%)
Query 9 VQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLS--NCTLSNDSEARVSC 66
V+K I+ G PG+GKGT + R+ H+S GDL+R++ S N E +
Sbjct 60 VEKPKIVFVLGGPGAGKGTQCSRIVDRFQFTHLSAGDLLREERSREGSEFGNLIEDYIRN 119
Query 67 GLLIQDTLASEICKDYISSHPERQSWILDGFPRTLQQVELLRGFATPHACINFIV----P 122
G ++ + + ++ + + + +++DGFPR ++ + F++
Sbjct 120 GKIVPVEVTCSLLENAMKASG-KSRFLIDGFPRNQDNLDGWNRQMSEKVDFQFVLFFDCG 178
Query 123 RHILLKKLLGR 133
+ +K+ LGR
Sbjct 179 EDVCVKRCLGR 189
> At3g01820
Length=219
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 53/112 (47%), Gaps = 3/112 (2%)
Query 11 KGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLS-NCTLSNDSEARVSCGLL 69
+G+ + G PG+ + + + L++ + H+S+G L+RQ+L+ +L + + V+ L
Sbjct 61 RGVQWVLMGAPGAWRHVFAERLSKLLEVPHISMGSLVRQELNPRSSLYKEIASAVNERKL 120
Query 70 IQDTLASEICKDYISSHPER--QSWILDGFPRTLQQVELLRGFATPHACINF 119
+ ++ + + R +IL G PRT Q E L A +N
Sbjct 121 VPKSVVFALLSKRLEEGYARGETGFILHGIPRTRFQAETLDQIAQIDLVVNL 172
> At5g60340_1
Length=196
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 45/95 (47%), Gaps = 5/95 (5%)
Query 15 LIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEARVSCGLLIQDTL 74
L+ +G PG+GK T LA +R++ +GDL+++K N+ E C + +D++
Sbjct 16 LLITGTPGTGKSTTASALAEATNLRYICIGDLVKEKEFYHGWDNELE----CHFINEDSV 71
Query 75 ASEICKDYISSHPERQSWILDGFP-RTLQQVELLR 108
E+ I D FP R +V +LR
Sbjct 72 IDELDDAMIEGGNIVDYHGCDFFPQRWFDRVVVLR 106
> Hs18582975
Length=723
Score = 38.1 bits (87), Expect = 0.012, Method: Composition-based stats.
Identities = 32/120 (26%), Positives = 45/120 (37%), Gaps = 31/120 (25%)
Query 19 GPPGSGKGTYGKILARRWGMRHVSLGDLIRQ---KLSNCTLSND---------------- 59
GPP GK + K LA + + H+ L D+I + KL ND
Sbjct 374 GPPAVGKSSIAKELANYYKLHHIQLKDVISEAIAKLEAIVAPNDVGEGEEEVEEEEEEEN 433
Query 60 -----------SEARVSCGLLIQDTLASEICKDYISSHPER-QSWILDGFPRTLQQVELL 107
E+ + D K+ + S P R Q +ILDGFP+T Q + L
Sbjct 434 VEDAQELLDGIKESMEQNAGQLDDQYIIRFMKEKLKSMPCRNQGYILDGFPKTYDQAKDL 493
> Hs21450759
Length=421
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 34/147 (23%), Positives = 68/147 (46%), Gaps = 12/147 (8%)
Query 9 VQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDS-EARVSCG 67
+ K + + G PG GK T + + + W V ++ ++++ T S ++ + G
Sbjct 27 LSKPVCFVVFGKPGVGKTTLARYITQAWKCIRVEALPILEEQIAAETESGVMLQSMLISG 86
Query 68 LLIQDTLASEICKDYISSHPE--RQSWILDGFPR-------TLQQVELLRGFA-TPHACI 117
I D L ++ + ++S PE +I+ P TLQQ+EL++ P I
Sbjct 87 QSIPDELVIKLMLEKLNS-PEVCHFGYIITEIPSLSQDAMTTLQQIELIKNLNLKPDVII 145
Query 118 NFIVPRHILLKKLLGRRICSSCGGCFN 144
N P + L +++ G+R ++ G ++
Sbjct 146 NIKCPDYDLCQRISGQRQHNNTGYIYS 172
> 7297473
Length=562
Score = 37.7 bits (86), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 44/98 (44%), Gaps = 6/98 (6%)
Query 13 IILIFSGPPGSGKGTY-GKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEARVSCGLLIQ 71
II + GP GS K T K + G H+S+G L+R + +N V L
Sbjct 275 IIWVIGGP-GSNKATLCLKAVGLNPGWAHISVGRLLRNITDSAPRANTESFAVKEALAAG 333
Query 72 DTLASEICKDYISSH----PERQSWILDGFPRTLQQVE 105
D + + ++ +R I+DG+PR LQQV+
Sbjct 334 DMAPEKSLNQLLETNLRQLRDRTGIIVDGYPRNLQQVK 371
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 21/93 (22%), Positives = 47/93 (50%), Gaps = 10/93 (10%)
Query 13 IILIFSGPPGSGKGTY-GKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEARVSCGLLIQ 71
+I + GP GSGK T+ + R G+ H+++ DL++Q + + S+ +
Sbjct 16 VIFVLGGP-GSGKVTHCDTFMQERRGVTHINMMDLLQQYAMGNDMQDFSQ--------LS 66
Query 72 DTLASEICKDYISSHPERQSWILDGFPRTLQQV 104
+E+ + P +++++ G+PR+++ V
Sbjct 67 SKTVTEVLMLEMKMAPAAKAYLISGYPRSMRDV 99
> SPCC1795.05c
Length=191
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 29/126 (23%), Positives = 58/126 (46%), Gaps = 6/126 (4%)
Query 14 ILIFSGPPGSGKGTYGKILARRWG-MRHVSLGDLIRQKLS--NCTLSNDSEARVSCGLLI 70
++ G PG+GKGT LA ++ H+S GD +R++ + N + + G ++
Sbjct 4 VIFVLGGPGAGKGTQCDRLAEKFDKFVHISAGDCLREEQNRPGSKYGNLIKEYIKDGKIV 63
Query 71 QDTLASEICKDYISSHPER--QSWILDGFPRTLQQVELLRGFATPHA-CINFIVPRHILL 127
+ + + + ++ +++DGFPR + Q E P + F + +L
Sbjct 64 PMEITISLLETKMKECHDKGIDKFLIDGFPREMDQCEGFEKSVCPAKFALYFRCGQETML 123
Query 128 KKLLGR 133
K+L+ R
Sbjct 124 KRLIHR 129
> ECU09g1330
Length=309
Score = 36.6 bits (83), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 15/21 (71%), Positives = 18/21 (85%), Gaps = 0/21 (0%)
Query 15 LIFSGPPGSGKGTYGKILARR 35
L+F+GPPG+GK T KILARR
Sbjct 38 LLFTGPPGTGKTTCAKILARR 58
> CE28286
Length=182
Score = 36.2 bits (82), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 33/64 (51%), Gaps = 4/64 (6%)
Query 15 LIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEARVSCGLLIQDTL 74
++ +G PG+GK T G+ +A + G + + +R+ L D + + +C +L +D L
Sbjct 12 ILVTGSPGTGKSTLGQQVAEKLGFVFIEVSKEVREN----NLQGDFDEQYNCHVLDEDKL 67
Query 75 ASEI 78
I
Sbjct 68 LDHI 71
> CE23016
Length=773
Score = 36.2 bits (82), Expect = 0.042, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query 11 KGIILIFSGPPGSGKGTYGKILARRWG--MRHVSLGDL 46
KG+IL F+GPPG GK + K +A G + VSLG +
Sbjct 328 KGMILCFTGPPGIGKTSIAKAIAESMGRKFQRVSLGGI 365
> SPAC1834.02
Length=1573
Score = 35.4 bits (80), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 39/86 (45%), Gaps = 4/86 (4%)
Query 3 EPLSTCVQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLSNDSEA 62
+PL + K +I G G+GK T GKI+A++ + + L +L+ L +E
Sbjct 854 DPLKGSISKNASIILIGMRGAGKTTIGKIIAKQLNFKFLDLDELLEDYLEMPI----AEV 909
Query 63 RVSCGLLIQDTLASEICKDYISSHPE 88
G ++ + +I+ HPE
Sbjct 910 IFRMGWDAFRLEEHKVLRKFITEHPE 935
> Hs19718790
Length=584
Score = 33.1 bits (74), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 10 QKGIILIFSGPPGSGKGTYGKILARRWGMR 39
Q G IL+ +GPPG GK T KIL++ G++
Sbjct 31 QGGSILLITGPPGCGKTTTLKILSKEHGIQ 60
> Hs4506383
Length=670
Score = 33.1 bits (74), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 10 QKGIILIFSGPPGSGKGTYGKILARRWGMR 39
Q G IL+ +GPPG GK T KIL++ G++
Sbjct 117 QGGSILLITGPPGCGKTTTLKILSKEHGIQ 146
> Hs19718786
Length=681
Score = 33.1 bits (74), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 10 QKGIILIFSGPPGSGKGTYGKILARRWGMR 39
Q G IL+ +GPPG GK T KIL++ G++
Sbjct 128 QGGSILLITGPPGCGKTTTLKILSKEHGIQ 157
> CE08422
Length=982
Score = 32.3 bits (72), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 14/89 (15%)
Query 99 RTLQQVELLRGFATPHACINFIVPRH----ILLKKLLGRRICSSCGGCFNVANICDEPYD 154
+ L + L+ GF A +N ++PRH I L +LG +C S N+C E
Sbjct 660 KVLWEHYLMHGFLDTQAVVNLLMPRHNCMVIRLTYILGGSVCVS------YRNLCGEIIH 713
Query 155 MPPILPTKECRHCKGNAPLTTRSDDTEEA 183
+ P+ E + + + L SD E A
Sbjct 714 LEPL----ELKKLQAKSILEYISDIAETA 738
> Hs14719833
Length=1443
Score = 32.0 bits (71), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 54/133 (40%), Gaps = 14/133 (10%)
Query 9 VQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLS-NCTLSNDSEARVSCG 67
Q+ I+I G G GK T + + WG R GD + +C S+
Sbjct 324 TQEPRIVILQGAAGIGKSTLARQVKEAWG-RGQLYGDRFQHVFYFSCRELAQSKVVSLAE 382
Query 68 LLIQDTLASEICKDYISSHPERQSWILDGFPR---TLQQ--VELLRGFATPHACINFIVP 122
L+ +D A+ I S PER +ILDG LQ+ EL ++ P P
Sbjct 383 LIGKDGTATPAPIRQILSRPERLLFILDGVDEPGWVLQEPSSELCLHWSQPQ-------P 435
Query 123 RHILLKKLLGRRI 135
LL LLG+ I
Sbjct 436 ADALLGSLLGKTI 448
> At5g19410
Length=624
Score = 32.0 bits (71), Expect = 0.83, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 6/75 (8%)
Query 2 HEPL----STCVQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLSNCTLS 57
H P+ S + IL GP G+GK T KI++ R + H +L ++N ++
Sbjct 63 HTPILNSVSLAAESSKILAVVGPSGTGKSTLLKIISGR--VNHKALDPSSAVLMNNRKIT 120
Query 58 NDSEARVSCGLLIQD 72
+ ++ R CG + QD
Sbjct 121 DYNQLRRLCGFVPQD 135
> Hs14719835
Length=1399
Score = 32.0 bits (71), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 54/133 (40%), Gaps = 14/133 (10%)
Query 9 VQKGIILIFSGPPGSGKGTYGKILARRWGMRHVSLGDLIRQKLS-NCTLSNDSEARVSCG 67
Q+ I+I G G GK T + + WG R GD + +C S+
Sbjct 324 TQEPRIVILQGAAGIGKSTLARQVKEAWG-RGQLYGDRFQHVFYFSCRELAQSKVVSLAE 382
Query 68 LLIQDTLASEICKDYISSHPERQSWILDGFPR---TLQQ--VELLRGFATPHACINFIVP 122
L+ +D A+ I S PER +ILDG LQ+ EL ++ P P
Sbjct 383 LIGKDGTATPAPIRQILSRPERLLFILDGVDEPGWVLQEPSSELCLHWSQPQ-------P 435
Query 123 RHILLKKLLGRRI 135
LL LLG+ I
Sbjct 436 ADALLGSLLGKTI 448
Lambda K H
0.323 0.140 0.448
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3443493696
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40