bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4267_orf2
Length=62
Score E
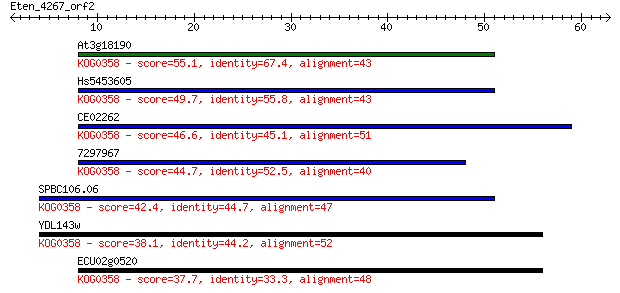
Sequences producing significant alignments: (Bits) Value
At3g18190 55.1 3e-08
Hs5453605 49.7 1e-06
CE02262 46.6 1e-05
7297967 44.7 4e-05
SPBC106.06 42.4 2e-04
YDL143w 38.1 0.004
ECU02g0520 37.7 0.006
> At3g18190
Length=536
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 29/43 (67%), Positives = 32/43 (74%), Gaps = 0/43 (0%)
Query 8 RMLREDRLLIAKMGKHIAATACICLLIQKSILRDAVTDLSLDY 50
R+L+E+R I M K I AT C LLIQKSILRDAVTDLSL Y
Sbjct 272 RILKEERNYILGMIKKIKATGCNVLLIQKSILRDAVTDLSLHY 314
> Hs5453605
Length=539
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 31/43 (72%), Gaps = 0/43 (0%)
Query 8 RMLREDRLLIAKMGKHIAATACICLLIQKSILRDAVTDLSLDY 50
R+LRE+R I + K I T C LLIQKSILRDA++DL+L +
Sbjct 274 RVLREERAYILNLVKQIKKTGCNVLLIQKSILRDALSDLALHF 316
> CE02262
Length=540
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 23/51 (45%), Positives = 32/51 (62%), Gaps = 0/51 (0%)
Query 8 RMLREDRLLIAKMGKHIAATACICLLIQKSILRDAVTDLSLDYRGESNNTC 58
R L+E+R + ++ K I A C LLIQKSILRDAV +L+L + + C
Sbjct 274 RALKEERQYLLEICKQIKAAGCNVLLIQKSILRDAVNELALHFLAKMKIMC 324
> 7297967
Length=533
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 21/40 (52%), Positives = 29/40 (72%), Gaps = 0/40 (0%)
Query 8 RMLREDRLLIAKMGKHIAATACICLLIQKSILRDAVTDLS 47
R+L+E+R I + K I + C LL+QKSILRDAV+DL+
Sbjct 268 RVLKEERSYILNIVKQIKKSGCNVLLVQKSILRDAVSDLA 307
> SPBC106.06
Length=527
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 4 RKWHRMLREDRLLIAKMGKHIAATACICLLIQKSILRDAVTDLSLDY 50
R+ ++L+E+R + M K I +LIQKSILRDAV DL+L +
Sbjct 259 RQMDKILKEERQYLLNMCKKIKKAGANVILIQKSILRDAVNDLALHF 305
> YDL143w
Length=528
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 23/52 (44%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 4 RKWHRMLREDRLLIAKMGKHIAATACICLLIQKSILRDAVTDLSLDYRGESN 55
R+ ++L+E+R + + K I C LLIQKSILRDAV DL+L + + N
Sbjct 258 RQMDKILKEERAYLLNICKKIKKAKCNVLLIQKSILRDAVNDLALHFLSKLN 309
> ECU02g0520
Length=484
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 8 RMLREDRLLIAKMGKHIAATACICLLIQKSILRDAVTDLSLDYRGESN 55
++++++R I +M K I + C L++QKSILR++++DL+ + + N
Sbjct 233 KIIQDERKYILEMCKKIKKSGCTLLVVQKSILRESLSDLASHFLKQLN 280
Lambda K H
0.328 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1175803722
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40