bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4260_orf1
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
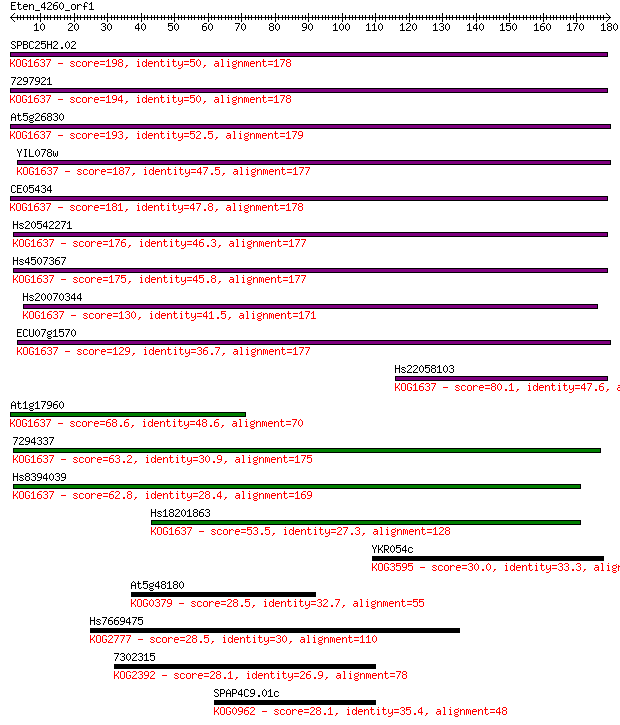
SPBC25H2.02 198 5e-51
7297921 194 1e-49
At5g26830 193 1e-49
YIL078w 187 9e-48
CE05434 181 8e-46
Hs20542271 176 2e-44
Hs4507367 175 4e-44
Hs20070344 130 2e-30
ECU07g1570 129 3e-30
Hs22058103 80.1 2e-15
At1g17960 68.6 7e-12
7294337 63.2 3e-10
Hs8394039 62.8 4e-10
Hs18201863 53.5 2e-07
YKR054c 30.0 2.7
At5g48180 28.5 7.2
Hs7669475 28.5 7.8
7302315 28.1 9.0
SPAP4C9.01c 28.1 9.9
> SPBC25H2.02
Length=703
Score = 198 bits (504), Expect = 5e-51, Method: Compositional matrix adjust.
Identities = 89/179 (49%), Positives = 125/179 (69%), Gaps = 1/179 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQ-KS 59
SC L++ F PE + VFWHSSAHILG+A E F L +GP GF+Y+ + +
Sbjct 101 ASCTLKLCDFNDPEGKRVFWHSSAHILGEATELSFHCHLCIGPPTDEGFFYEMGIDNGRV 160
Query 60 LSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVY 119
++ ++ ++ + A++ + + QPFERLV +KE LEMF N +K +I TK+PDG TTVY
Sbjct 161 ITNDDYSSIESYAKQAIKQKQPFERLVISKEGLLEMFRYNKYKQYIIQTKIPDGASTTVY 220
Query 120 RCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
RCG L+D+C GPHVP T KSF+VT++S++Y+LG+ NDSLQRVYG+SFP+ KQ+ EY
Sbjct 221 RCGPLIDLCTGPHVPHTGRIKSFAVTKNSSSYFLGDAKNDSLQRVYGISFPDNKQMQEY 279
> 7297921
Length=690
Score = 194 bits (492), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 89/178 (50%), Positives = 123/178 (69%), Gaps = 1/178 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
G+C LQ+LKF PEAQ VFWHSSAHI+G+AME +G L GP ++ GFYYD ++ + +
Sbjct 104 GNCTLQLLKFDDPEAQAVFWHSSAHIMGEAMERIYGGHLCYGPPIENGFYYDMHLEGEGI 163
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
S +++GA+ ++IV E Q FERL K + LEMF N FKV ++ KV TTVY+
Sbjct 164 STNDYGAMEGLVKQIVKEKQNFERLEMKKSDLLEMFKYNEFKVRILNEKVT-TDRTTVYK 222
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
CG+L+D+CRGPHV T K+ +T++S+ YW G ++LQRVYG+SFP+ KQL E+
Sbjct 223 CGSLIDLCRGPHVRHTGKVKALKITKNSSTYWEGKADAETLQRVYGISFPDPKQLKEW 280
> At5g26830
Length=676
Score = 193 bits (491), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 94/181 (51%), Positives = 127/181 (70%), Gaps = 3/181 (1%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGP--ALKPGFYYDCYMGQK 58
G CKL++ KF S + ++ WHSSAHILGQA+E E+G L +GP GFYYD + G+
Sbjct 94 GDCKLELFKFDSDKGRDTLWHSSAHILGQALEQEYGCQLCIGPCTTRGEGFYYDGFYGEL 153
Query 59 SLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTV 118
LS++ ++ A A + EAQPFER+ TK++ALEMF+EN FKV LI +P TV
Sbjct 154 GLSDNHFPSIEAGAAKAAKEAQPFERIEVTKDQALEMFSENNFKVELING-LPADMTITV 212
Query 119 YRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
YRCG LVD+CRGPH+P T+ K+F R S+AYW G++ +SLQRVYG+S+P++KQL +Y
Sbjct 213 YRCGPLVDLCRGPHIPNTSFVKAFKCLRASSAYWKGDKDRESLQRVYGISYPDQKQLKKY 272
Query 179 L 179
L
Sbjct 273 L 273
> YIL078w
Length=734
Score = 187 bits (475), Expect = 9e-48, Method: Compositional matrix adjust.
Identities = 84/187 (44%), Positives = 127/187 (67%), Gaps = 10/187 (5%)
Query 3 CKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMG------ 56
KL++L F S E + VFWHSSAH+LG++ EC GA + +GP GF+Y+ +
Sbjct 133 IKLELLDFESDEGKKVFWHSSAHVLGESCECHLGAHICLGPPTDDGFFYEMAVRDSMKDI 192
Query 57 ----QKSLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPD 112
++++S+++ L A+ ++ + Q FERLV +KE+ L+MF + +K L+ TKVPD
Sbjct 193 SESPERTVSQADFPGLEGVAKNVIKQKQKFERLVMSKEDLLKMFHYSKYKTYLVQTKVPD 252
Query 113 GGLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEK 172
GG TTVYRCG L+D+C GPH+P T K+F + ++S+ Y+LG+ +NDSLQRVYG+SFP+K
Sbjct 253 GGATTVYRCGKLIDLCVGPHIPHTGRIKAFKLLKNSSCYFLGDATNDSLQRVYGISFPDK 312
Query 173 KQLTEYL 179
K + +L
Sbjct 313 KLMDAHL 319
> CE05434
Length=725
Score = 181 bits (458), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 85/178 (47%), Positives = 119/178 (66%), Gaps = 1/178 (0%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSL 60
G+ KL++LKF EA+ VFWHSSAH+LG+AME G L GP ++ GFYYD + +++
Sbjct 134 GNAKLELLKFDDDEAKQVFWHSSAHVLGEAMERYCGGHLCYGPPIQEGFYYDMWHENRTI 193
Query 61 SESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYR 120
+ ++ + V + Q FERL TKE+ LEMF N FKV +IT K+ TTVYR
Sbjct 194 CPDDFPKIDQIVKAAVKDKQKFERLEMTKEDLLEMFKYNEFKVRIITEKIHTPK-TTVYR 252
Query 121 CGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
CG L+D+CRGPHV T K+ ++T++S++YW G +SLQR+YG+SFP+ KQL E+
Sbjct 253 CGPLIDLCRGPHVRHTGKVKAMAITKNSSSYWEGKADAESLQRLYGISFPDSKQLKEW 310
> Hs20542271
Length=723
Score = 176 bits (447), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 82/177 (46%), Positives = 115/177 (64%), Gaps = 1/177 (0%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
C L++LKF EAQ V+WHSSAHI+G+AME +G L GP ++ GFYYD Y+ + +S
Sbjct 136 DCTLELLKFEDEEAQAVYWHSSAHIMGEAMERVYGGCLCYGPPIENGFYYDMYLEEGGVS 195
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
++ +L A ++I+ E Q FERL KE L MF N FK ++ KV + TTVYRC
Sbjct 196 SNDFSSLEALCKKIIKEKQAFERLEVKKETLLAMFKYNKFKCRILNEKV-NTPTTTVYRC 254
Query 122 GTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
G L+D+CRGPHV T K+ + ++S+ YW G ++LQR+YG+SFP+ K L E+
Sbjct 255 GPLIDLCRGPHVRHTGKIKALKIHKNSSTYWEGKADMETLQRIYGISFPDPKMLKEW 311
> Hs4507367
Length=712
Score = 175 bits (444), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 81/177 (45%), Positives = 114/177 (64%), Gaps = 1/177 (0%)
Query 2 SCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLS 61
C L++LKF EAQ V+WHSSAHI+G+ ME +G L GP ++ GFYYD Y+ + +S
Sbjct 124 DCTLELLKFEDEEAQAVYWHSSAHIMGEGMERVYGGCLCYGPPIENGFYYDMYLEEGGVS 183
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
++ +L A ++I+ E Q FERL KE L MF N FK ++ KV + TTVYRC
Sbjct 184 SNDFSSLEALCKKIIKEKQAFERLEVKKETLLAMFKYNKFKCRILNEKV-NTPTTTVYRC 242
Query 122 GTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEY 178
G L+D+CRGPHV T K+ + ++S+ YW G ++LQR+YG+SFP+ K L E+
Sbjct 243 GPLIDLCRGPHVRHTGKIKALKIHKNSSTYWEGKADMETLQRIYGISFPDPKMLKEW 299
> Hs20070344
Length=718
Score = 130 bits (326), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 71/172 (41%), Positives = 108/172 (62%), Gaps = 2/172 (1%)
Query 5 LQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGPALKPGFYYDCYMG-QKSLSES 63
L+ L F SPE + VFWHSS H+LG A E GA+L GP+ + GFY+D ++G ++++ S
Sbjct 117 LRFLTFDSPEGKAVFWHSSTHVLGAAAEQFLGAVLCRGPSTEYGFYHDFFLGKERTIRGS 176
Query 64 EHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRCGT 123
E L Q + + A+PF RL ++++ ++F +NPFK+ LI KV G TVY CGT
Sbjct 177 ELPVLERICQELTAAARPFRRLEASRDQLRQLFKDNPFKLHLIEEKV-TGPTATVYGCGT 235
Query 124 LVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQL 175
LVD+C+GPH+ T + +S++ W + + ++LQRV G+SFP + L
Sbjct 236 LVDLCQGPHLRHTGQIGGLKLLSNSSSLWRSSGAPETLQRVSGISFPTTELL 287
> ECU07g1570
Length=640
Score = 129 bits (325), Expect = 3e-30, Method: Composition-based stats.
Identities = 65/178 (36%), Positives = 107/178 (60%), Gaps = 4/178 (2%)
Query 3 CKLQILKFVSPEAQNVFWHSSAHILGQAMECEF-GALLTVGPALKPGFYYDCYMGQKSLS 61
CKL+++ F A+++FWHSSAH+LG A+ + A L GP ++ GF+YD + +S
Sbjct 57 CKLELMTFEDDGARDIFWHSSAHVLGNALVNLYPDAKLVHGPPIEEGFFYDVDV-TDPIS 115
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALITTKVPDGGLTTVYRC 121
++ + +I+ + FER++ +KEE LE + +NP K I V ++VYR
Sbjct 116 SEDYEKIEEEMVKIMKKNYRFERVIKSKEELLEAYRDNPCKTHFIMKGVDKE--SSVYRN 173
Query 122 GTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQLTEYL 179
G D+C GPH+ +T + K+ V ++S+AY+L + + SLQR+Y ++FP K + EYL
Sbjct 174 GDFFDMCLGPHIRSTGVIKAVKVLKNSSAYFLNDPNLKSLQRIYAITFPSKGMMDEYL 231
> Hs22058103
Length=707
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 30/63 (47%), Positives = 46/63 (73%), Gaps = 0/63 (0%)
Query 116 TTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVSFPEKKQL 175
TTVYRCG L+D+C+GPHV T K+ + ++S+ YW GN ++LQR+YG+SFP+ K +
Sbjct 233 TTVYRCGPLIDLCKGPHVRHTGKIKTIKIFKNSSTYWEGNPEMETLQRIYGISFPDNKMM 292
Query 176 TEY 178
++
Sbjct 293 RDW 295
> At1g17960
Length=458
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 45/73 (61%), Gaps = 3/73 (4%)
Query 1 GSCKLQILKFVSPEAQNVFWHSSAHILGQAMECEFGALLTVGP--ALKPGFYYDC-YMGQ 57
G C L+I F S + +N FWHSSAHILGQA+E E+G L +GP GFYYD Y G+
Sbjct 96 GDCSLEIFGFDSDQGRNTFWHSSAHILGQALEQEYGCKLCIGPCEPRDEGFYYDSLYYGE 155
Query 58 KSLSESEHGALNA 70
L+++ + A
Sbjct 156 LGLNDNHFPNIEA 168
> 7294337
Length=341
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 54/189 (28%), Positives = 88/189 (46%), Gaps = 17/189 (8%)
Query 2 SCKLQILKFVSPEAQNV---FWHSSAHILGQAMECEFGALLTV------GPALKPG-FYY 51
SC LQ+L F E V FW + + +LG A+ F + GP +K G F +
Sbjct 115 SCTLQLLNFHVSEPHVVNKAFWRTCSFMLGAALNRAFKPEANLQLHSFPGPNIKSGSFVH 174
Query 52 DCYMGQKSLS--ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKV-ALITT 108
D + ++ + E AL+A ++ ++ ERL ++ A EMF ++ +K L +
Sbjct 175 DIVLQTQNWEPGKEEMRALSAEMVKLAAQDLRIERLDVQQDLAQEMFKDSKYKSEQLPSI 234
Query 109 KVPDGGLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRS-NDSLQRVYGV 167
G T+YR G +DI RGP V +T+ + SAA+ + + + R+ GV
Sbjct 235 SQQTNGRVTLYRLGDHIDISRGPMVASTSFLGKCVI---SAAHKVAEEGPSGAFYRIQGV 291
Query 168 SFPEKKQLT 176
+ P QL
Sbjct 292 ALPSGFQLN 300
> Hs8394039
Length=258
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 48/185 (25%), Positives = 79/185 (42%), Gaps = 19/185 (10%)
Query 2 SCKLQILKF--VSP-EAQNVFWHSSAHILGQAMECEFGALLTVG-------PALKPGFYY 51
SC+++ L F P E +W S A ++G +E F V P + F Y
Sbjct 42 SCEIKFLTFKDCDPGEVNEAYWRSCAMMMGCVIERAFKDEYMVNLVRAPEVPVISGAFCY 101
Query 52 DCYMGQK----SLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAENPFKVALIT 107
D + K ++ + A ++ + PFE L + ALE+F + +KV I
Sbjct 102 DVVLDSKLDEWMPTKENLRSFTKDAHALIYKDLPFETLEVEAKVALEIFQHSKYKVDFIE 161
Query 108 TKVPDG--GLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVY 165
K + ++R G +D+ GP +P T++C + V SA + L ++R
Sbjct 162 EKASQNPERIVKLHRIGDFIDVSEGPLIPRTSICFQYEV---SAVHNLQPTQPSLIRRFQ 218
Query 166 GVSFP 170
GVS P
Sbjct 219 GVSLP 223
> Hs18201863
Length=204
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/134 (26%), Positives = 59/134 (44%), Gaps = 9/134 (6%)
Query 43 PALKPGFYYDCYMGQK----SLSESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMFAE 98
P + F YD + K ++ + A ++ + PFE L + ALE+F
Sbjct 24 PVISGAFCYDVVLDSKLDEWMPTKENLRSFTKDAHALIYKDLPFETLEVEAKVALEIFQH 83
Query 99 NPFKVALITTKVPDG--GLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNR 156
+ +KV I K + ++R G +D+ GP +P T++C + V SA + L
Sbjct 84 SKYKVDFIEEKASQNPERIVKLHRIGDFIDVSEGPLIPRTSICFQYEV---SAVHNLQPT 140
Query 157 SNDSLQRVYGVSFP 170
++R GVS P
Sbjct 141 QPSLIRRFQGVSLP 154
> YKR054c
Length=4092
Score = 30.0 bits (66), Expect = 2.7, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 9/76 (11%)
Query 109 KVPDGGLTTVYRCGTLVDICRGPHVPTTALCKSFSVTRHSAAYWLGNRSNDSLQRVYGVS 168
K P+ T+ R +V C P P + S TRH+A +LG S SL ++Y +
Sbjct 2517 KTPENKWVTIERI-HIVGACNPPTDPGR-IPMSERFTRHAAILYLGYPSGKSLSQIYEIY 2574
Query 169 F-------PEKKQLTE 177
+ PE + TE
Sbjct 2575 YKAIFKLVPEFRSYTE 2590
> At5g48180
Length=301
Score = 28.5 bits (62), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 3/58 (5%)
Query 37 ALLTVGPALKPGFYYDCYMGQKSLSESEH---GALNAAAQRIVSEAQPFERLVCTKEE 91
A +V PA+ G Y Y G++ E H G ++ ++ +E +ER+VC EE
Sbjct 195 AARSVFPAVSYGKYIVIYGGEEEPHELMHMGAGKMSGEVYQLDTETLVWERIVCGNEE 252
> Hs7669475
Length=1181
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 51/122 (41%), Gaps = 20/122 (16%)
Query 25 HILGQAMECEFGALLTVGPALKPGFYYDCYMGQKSLSESEHGALNAAAQRIVSEAQPFER 84
H LG + CEF L GPA +P F Y +G ++ +++A ++++ + E
Sbjct 624 HKLGNS--CEFRLLSKEGPAHEPKFQYCVAVGAQTFP-----SVSAPSKKVAKQMAAEEA 676
Query 85 LVCTKEEALE-MFAENPFK-----VALITTKVPDGGLTTVYRCG------TLVDICRGPH 132
+ EA M ++N + V + T P GGL R LVD PH
Sbjct 677 MKALHGEATNSMASDNQVRKIGELVRYLNTN-PVGGLLEYARSHGFAAEFKLVDQSGPPH 735
Query 133 VP 134
P
Sbjct 736 EP 737
> 7302315
Length=431
Score = 28.1 bits (61), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 35/78 (44%), Gaps = 7/78 (8%)
Query 32 ECEFGALLTVGPALKPGFYYDCYMGQKSLSESEHGALNAAAQRIVSEAQPFERLVCTKEE 91
+ EFG +L LK + + + +E G AAA + + R + + EE
Sbjct 336 QAEFGKMLQSPEPLK----VSAIIHKAFIEVNEEGTEAAAATGMAVRRK---RAIMSPEE 388
Query 92 ALEMFAENPFKVALITTK 109
+E FA++PF L+ K
Sbjct 389 PIEFFADHPFTYVLVHQK 406
> SPAP4C9.01c
Length=666
Score = 28.1 bits (61), Expect = 9.9, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Query 62 ESEHGALNAAAQRIVSEAQPFERLVCTKEEALEMF---AENPFKVALITTK 109
+S++GALNA ++ E + E + +E L M A+ F+ LI TK
Sbjct 424 KSKYGALNAERAGLLGECKQLENSITKDKEELNMEFKDADERFRRQLIKTK 474
Lambda K H
0.320 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2815454148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40