bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4233_orf4
Length=233
Score E
Sequences producing significant alignments: (Bits) Value
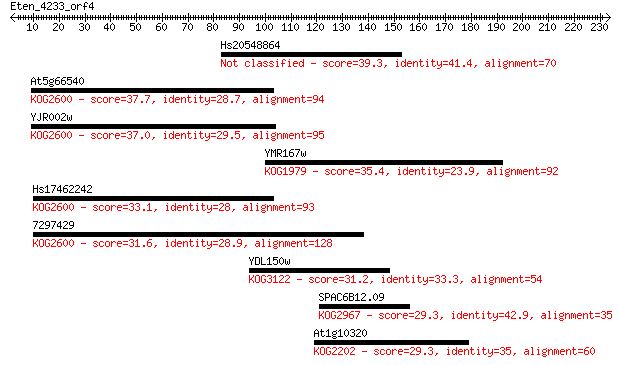
Hs20548864 39.3 0.007
At5g66540 37.7 0.018
YJR002w 37.0 0.038
YMR167w 35.4 0.095
Hs17462242 33.1 0.46
7297429 31.6 1.5
YDL150w 31.2 1.8
SPAC6B12.09 29.3 6.9
At1g10320 29.3 7.2
> Hs20548864
Length=556
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 38/73 (52%), Gaps = 15/73 (20%)
Query 83 DAPASVRVEEAIPVYVPAASSSSSSSKAALPFTPKTAAEATREERRAARRSKKAKRQQ-- 140
+AP ++ +EEA SSSKA P T +T+A A R R R S+ RQQ
Sbjct 145 EAPRTLAIEEA------------SSSKAREPITSQTSAWALRSCREVTRHSRGICRQQES 192
Query 141 -QLRRRIDAGELT 152
QL R +D+GE T
Sbjct 193 KQLLRAVDSGEET 205
> At5g66540
Length=524
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 46/96 (47%), Gaps = 8/96 (8%)
Query 9 ELNMEKPAEGLGDIYAQQYREKV--LGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNY 66
EL+ K +GL ++Y +Y +K AP+ +L K++ LF ++ K DAL+++
Sbjct 379 ELDESKSKKGLAEVYEAEYFQKANPAFAPTTHSDEL---KKEASMLFKKLCLKLDALSHF 435
Query 67 QFVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAAS 102
F P+ A + +EE PV V A+
Sbjct 436 HFTPKPVIEEMSIPNVSA---IAMEEVAPVAVSDAA 468
> YJR002w
Length=593
Score = 37.0 bits (84), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 52/96 (54%), Gaps = 6/96 (6%)
Query 9 ELNMEKPAEGLGDIYAQQY-REKVLGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNYQ 67
EL+ K ++ L +IY Y R + A S++ Q+ E + +L++ ++YK D L++
Sbjct 399 ELSDVKSSKSLAEIYEDDYTRAEDESALSEELQKAHSE---ISELYANLVYKLDVLSSVH 455
Query 68 FVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASS 103
FVP+ + P ++ +E+A P+Y+ ASS
Sbjct 456 FVPK-PASTSLEIRVETP-TISMEDAQPLYMSNASS 489
> YMR167w
Length=769
Score = 35.4 bits (80), Expect = 0.095, Method: Composition-based stats.
Identities = 22/103 (21%), Positives = 48/103 (46%), Gaps = 11/103 (10%)
Query 100 AASSSSSSSKAALPFTPKTAAEATREERRAAR-----------RSKKAKRQQQLRRRIDA 148
A+S S++ ++ +PF ++ R+ R A+ + +KAKRQ+ RIDA
Sbjct 345 ASSISTNKPESLIPFNDTIESDRNRKSLRQAQVVENSYTTANSQLRKAKRQENKLVRIDA 404
Query 149 GELTLQQAQQRQQLLQQKNKAAKEEKKHKRLTGLTHAEALKEL 191
+ + Q + + K + ++T ++H++ ++L
Sbjct 405 SQAKITSFLSSSQQFNFEGSSTKRQLSEPKVTNVSHSQEAEKL 447
> Hs17462242
Length=681
Score = 33.1 bits (74), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 11/96 (11%)
Query 10 LNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQLLQ---LFSQIMYKCDALTNY 66
L+ EK L +IY Q+Y QQ+ ++E + ++ + + K DAL+N+
Sbjct 460 LDHEKSKLSLAEIYEQEY------IKLNQQKTAEEENPEHVEIQKMMDSLFLKLDALSNF 513
Query 67 QFVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAAS 102
F+P+ + PA + +EE PV V A+
Sbjct 514 HFIPK-PPVPEIKVVSNLPA-ITMEEVAPVSVSDAA 547
> 7297429
Length=662
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 37/134 (27%), Positives = 55/134 (41%), Gaps = 8/134 (5%)
Query 10 LNMEKPAEGLGDIYAQQY-REKVLGAPSQQQQQLD----QEKQQLLQLFSQIMYKCDALT 64
L+ EK + L IY QY RE P++ +E Q++ + + K DAL+
Sbjct 440 LDQEKSKQSLSQIYEAQYQREMEKLDPNRDADDKTGPEPKEHQEIKRAMRSLFLKLDALS 499
Query 65 NYQFVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASSSSSSSKAALP-FTPKTAAEAT 123
N+ F P+ + PA V +EE P+ V A + P P E
Sbjct 500 NFHFTPK-PVAPEIKIVTNTPA-VHMEEVAPLAVSDAKLLAPEEVFRGPKHAPLGKTERD 557
Query 124 REERRAARRSKKAK 137
R ++ RR KK K
Sbjct 558 RTDKNRERRKKKQK 571
> YDL150w
Length=422
Score = 31.2 bits (69), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 2/54 (3%)
Query 94 IPVYVPAASSSSSSSKAALPFTPKTAAEATREERRAARRSKKAKRQQQLRRRID 147
+P ++S+ S+K +L F PK A ++EER AA + K K +++ +R D
Sbjct 10 LPSLKDSSSNGGGSAKPSLKFKPKAVARKSKEEREAA--ASKVKLEEESKRGND 61
> SPAC6B12.09
Length=304
Score = 29.3 bits (64), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 121 EATREERRAARRSKKAKRQQQLRRRIDAGELTLQQ 155
+A RE+R RR KK R+++ +R+I+AGE+ Q
Sbjct 47 DAGREKRAEMRREKKRLRKEERKRKIEAGEVVKSQ 81
> At1g10320
Length=757
Score = 29.3 bits (64), Expect = 7.2, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 33/60 (55%), Gaps = 3/60 (5%)
Query 119 AAEATREERRAARRSKKAKRQQQLRRRIDAGELTLQQAQQRQQLLQQKNKAAKEEKKHKR 178
AAE +R+E+R A + K K Q+R+ I A E +A+ Q++ KA +EE +R
Sbjct 30 AAEMSRKEKRKAMKKLKRK---QVRKEIAAKEREEAKAKLNDPAEQERLKAIEEEDARRR 86
Lambda K H
0.314 0.125 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4562417408
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40