bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4218_orf1
Length=269
Score E
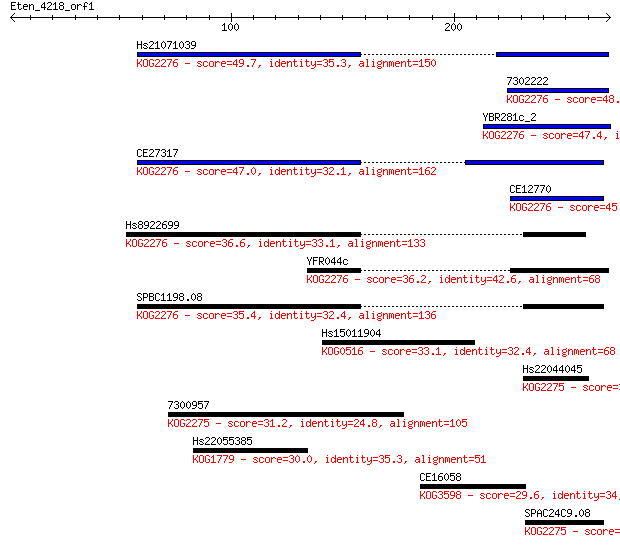
Sequences producing significant alignments: (Bits) Value
Hs21071039 49.7 7e-06
7302222 48.9 1e-05
YBR281c_2 47.4 3e-05
CE27317 47.0 4e-05
CE12770 45.8 9e-05
Hs8922699 36.6 0.059
YFR044c 36.2 0.083
SPBC1198.08 35.4 0.12
Hs15011904 33.1 0.68
Hs22044045 32.3 1.1
7300957 31.2 2.2
Hs22055385 30.0 5.1
CE16058 29.6 6.4
SPAC24C9.08 29.3 9.4
> Hs21071039
Length=507
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 219 GARSNSPGWHGTGWHTNPWKIHXQDGYIYGRGVTDNKGPIICSLLAVKHF 268
G P G GW T+P+ + DG +YGRG TDNKGP++ + AV F
Sbjct 131 GHLDVQPADRGDGWLTDPYVLTEVDGKLYGRGATDNKGPVLAWINAVSAF 180
Score = 32.3 bits (72), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 45/105 (42%), Gaps = 16/105 (15%)
Query 58 LEPRRRDFLPLLAQFVAYKSVSASRCPRHITGCIGAAKFVAQLLEQALGATVDIV--WPQ 115
++ + +F+ L ++VA +S S PR A L Q LGA V V PQ
Sbjct 42 IDLHQDEFVQTLKEWVAIESDSVQPVPRFRQELFRMMAVAADTL-QRLGARVASVDMGPQ 100
Query 116 RSAVLSEFHGDQWEGQPHP---VVFGRLGTNPHHPTIVFYSHYDV 157
+ +GQ P V+ LG++P T+ FY H DV
Sbjct 101 QLP----------DGQSLPIPPVILAELGSDPTKGTVCFYGHLDV 135
> 7302222
Length=462
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 31/45 (68%), Gaps = 0/45 (0%)
Query 224 SPGWHGTGWHTNPWKIHXQDGYIYGRGVTDNKGPIICSLLAVKHF 268
P GW+TNP+++ DG ++GRG +D+KGP++C + A++ +
Sbjct 107 QPALKEDGWNTNPFELTEVDGKLFGRGASDDKGPVLCWIHAIEAY 151
> YBR281c_2
Length=452
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 40/67 (59%), Gaps = 10/67 (14%)
Query 213 QIAKHRGARSNSPGWHG----------TGWHTNPWKIHXQDGYIYGRGVTDNKGPIICSL 262
++++ +GA+ W+G W+T+P+ + ++GY+ GRGV+DNKGP++ ++
Sbjct 77 KVSQVKGAKKKRILWYGHYDVISSGNTFNWNTDPFTLTCENGYLKGRGVSDNKGPLVSAI 136
Query 263 LAVKHFI 269
+V +
Sbjct 137 HSVAYLF 143
> CE27317
Length=461
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 33/62 (53%), Gaps = 3/62 (4%)
Query 205 RSSSNSVAQIAKHRGARSNSPGWHGTGWHTNPWKIHXQDGYIYGRGVTDNKGPIICSLLA 264
R S I H + P GW+TNP+++ DG ++GRG TD+KGP+I +
Sbjct 87 RDKSKKTLLIYGHLDVQ---PAEKEDGWNTNPFELTEIDGKLFGRGSTDDKGPVIAWIAV 143
Query 265 VK 266
+K
Sbjct 144 LK 145
Score = 33.1 bits (74), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 47/101 (46%), Gaps = 10/101 (9%)
Query 58 LEPRRRDFLPLLAQFVAYKSVSASRCPRHITGCIGAAKFVAQLLEQALGATVDIVWPQRS 117
++ R+ +F+ LL + VA +SVSA P C+ +++ L + LG + W
Sbjct 11 IDERQDEFIDLLRESVAIQSVSAD--PARRGDCVRMSEWARDQL-KTLGVETSL-WELGQ 66
Query 118 AVLSEFHGDQWEGQPHP-VVFGRLGTNPHHPTIVFYSHYDV 157
L G+Q P P VFG G + T++ Y H DV
Sbjct 67 QTLP--SGEQL---PLPPAVFGVYGRDKSKKTLLIYGHLDV 102
> CE12770
Length=473
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 225 PGWHGTGWHTNPWKIHXQDGYIYGRGVTDNKGPIICSLLAVK 266
P GW T P+++ +DG ++GRG +D+KGP++C A++
Sbjct 104 PAAKSDGWDTEPFELVEKDGKLFGRGSSDDKGPVLCWFHAIR 145
> Hs8922699
Length=475
Score = 36.6 bits (83), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 51/105 (48%), Gaps = 8/105 (7%)
Query 53 TMKTGLEPRRRDFLPLLAQFVAYKSVSASRCPRHITGCIGAAKFVAQLLEQALGATVDIV 112
T+ ++ + ++ LA++VA +SVSA R G I VA + LG +V++V
Sbjct 6 TLFKYIDENQDRYIKKLAKWVAIQSVSAWPEKR---GEIRRMMEVAAADVKQLGGSVELV 62
Query 113 WPQRSAVLSEFHGDQWEGQPHPVVFGRLGTNPHHPTIVFYSHYDV 157
+ + D E P++ GRLG++P T+ Y H DV
Sbjct 63 DIGKQKL-----PDGSEIPLPPILLGRLGSDPQKKTVCIYGHLDV 102
Score = 36.2 bits (82), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 231 GWHTNPWKIHXQDGYIYGRGVTDNKGPI 258
GW + P+ + +DG +YG G TD+KGP+
Sbjct 110 GWDSEPFTLVERDGKLYGGGSTDDKGPV 137
> YFR044c
Length=481
Score = 36.2 bits (82), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Query 225 PGWHGTGWHTNPWK--IHXQDGYIYGRGVTDNKGPIICSLLAVKHF 268
P GW T P+K I G + GRGVTD+ GP++ + V F
Sbjct 107 PAQLEDGWDTEPFKLVIDEAKGIMKGRGVTDDTGPLLSWINVVDAF 152
Score = 33.5 bits (75), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 134 PVVFGRLGTNPHHPTIVFYSHYDV 157
PV+ R G++P T++ Y HYDV
Sbjct 82 PVILSRFGSDPSKKTVLVYGHYDV 105
> SPBC1198.08
Length=474
Score = 35.4 bits (80), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 52/103 (50%), Gaps = 15/103 (14%)
Query 58 LEPRRRDFLPLLAQFVAYKSVSASRCPRHITGCIGAAKFVAQLLEQALGATVDIVWPQRS 117
++ ++ +F+ L++ V+ SVSA R + A FV + LGA ++ +R
Sbjct 10 IDKKKDEFVTRLSRAVSIPSVSADVTLR--PKVVEMADFVVSEFTK-LGAKME----KRD 62
Query 118 AVLSEFHGDQWEGQPHP---VVFGRLGTNPHHPTIVFYSHYDV 157
+H Q +GQ P +V G+ G +P T++ Y+H+DV
Sbjct 63 I---GYH--QMDGQDVPLPPIVLGQYGNDPSKKTVLIYNHFDV 100
Score = 35.4 bits (80), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 26/37 (70%), Gaps = 1/37 (2%)
Query 231 GWHTNPWKIHXQD-GYIYGRGVTDNKGPIICSLLAVK 266
GW T+P+ + + G ++GRG TD+KGP+I + A++
Sbjct 108 GWSTDPFTLTVDNKGRMFGRGATDDKGPLIGWISAIE 144
> Hs15011904
Length=5430
Score = 33.1 bits (74), Expect = 0.68, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 7/68 (10%)
Query 141 GTNPHHPTIVFYSHYDVVSAGGTSPSSVPASSDVSEGSASPVTVSASPRTRTRTVVPPSN 200
G+ PT F+S + AG TS SS PAS+ A P ++ P +R +
Sbjct 5308 GSKLKRPTPTFHSSRTSL-AGDTSNSSSPASTGAKTNRADPKKSASRPGSRA------GS 5360
Query 201 QQGARSSS 208
+ G+R+SS
Sbjct 5361 RAGSRASS 5368
> Hs22044045
Length=361
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 231 GWHTNPWKIHXQDGYIYGRGVTDNKGPII 259
GW P+ +DG IYGRG D+K ++
Sbjct 135 GWEVPPFSGLERDGIIYGRGTLDDKNSVM 163
> 7300957
Length=400
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 43/106 (40%), Gaps = 20/106 (18%)
Query 72 FVAYKSVSASRCPRHITGCIGAAKFVAQLLEQALGATVDIVWPQRSAVLSEFHGDQWEGQ 131
F Y ++ + T C+ K A +L VD+++P +
Sbjct 15 FREYLRIATVQPNVDYTECVEFLKRQAH----SLDLPVDVIYPV---------------E 55
Query 132 PHPVVFGR-LGTNPHHPTIVFYSHYDVVSAGGTSPSSVPASSDVSE 176
PVV + LGT P P+++ SH DVV + P ++D+ E
Sbjct 56 KKPVVIIKWLGTEPELPSVILNSHMDVVPVFRDKWTHDPFAADIDE 101
> Hs22055385
Length=232
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 24/53 (45%), Gaps = 2/53 (3%)
Query 83 CPRHITGCIG--AAKFVAQLLEQALGATVDIVWPQRSAVLSEFHGDQWEGQPH 133
CP +I G + KF A E+ + A DI+W R A E D W+ H
Sbjct 75 CPDNIGGDLTFRDHKFSAHFTEEEVQALEDILWEHRPANQLEGVVDTWQASTH 127
> CE16058
Length=3498
Score = 29.6 bits (65), Expect = 6.4, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 22/47 (46%), Gaps = 3/47 (6%)
Query 185 SASPRTRTRTVVPPSNQQGARSSSNSVAQIAKHRGARSNSPGWHGTG 231
S P+ T P N AR+ S ++ QIA G +N G+ TG
Sbjct 2996 SGRPQMHT---TPTKNDMSARAPSGAMGQIANRMGHGTNPQGYGSTG 3039
> SPAC24C9.08
Length=596
Score = 29.3 bits (64), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 10/35 (28%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 232 WHTNPWKIHXQDGYIYGRGVTDNKGPIICSLLAVK 266
W+ P+ +G++Y RG D+K ++ L A++
Sbjct 211 WYFPPFSATYHNGHVYSRGAADDKNSVVAILEALE 245
Lambda K H
0.317 0.132 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5737723452
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40