bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4186_orf2
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
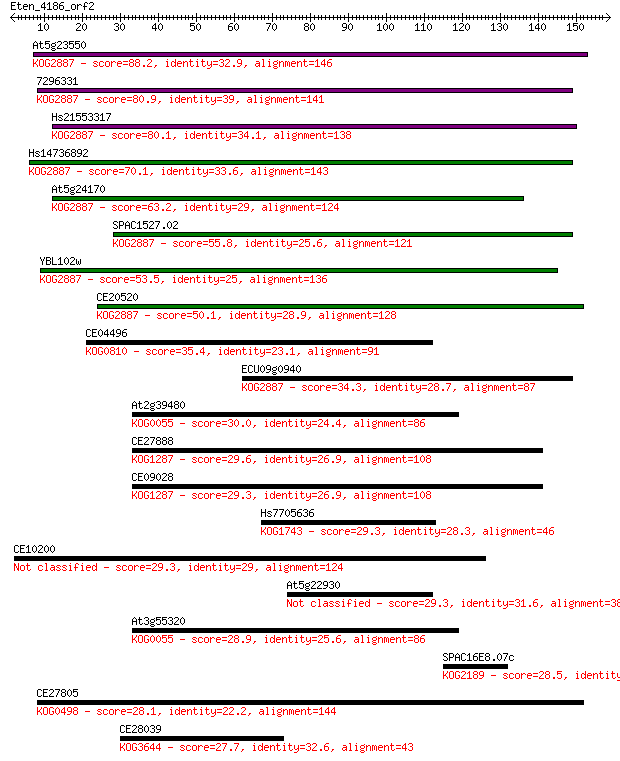
At5g23550 88.2 6e-18
7296331 80.9 1e-15
Hs21553317 80.1 1e-15
Hs14736892 70.1 2e-12
At5g24170 63.2 2e-10
SPAC1527.02 55.8 3e-08
YBL102w 53.5 2e-07
CE20520 50.1 2e-06
CE04496 35.4 0.042
ECU09g0940 34.3 0.11
At2g39480 30.0 1.8
CE27888 29.6 2.6
CE09028 29.3 2.9
Hs7705636 29.3 3.0
CE10200 29.3 3.5
At5g22930 29.3 3.5
At3g55320 28.9 4.3
SPAC16E8.07c 28.5 6.4
CE27805 28.1 7.2
CE28039 27.7 9.3
> At5g23550
Length=175
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 48/157 (30%), Positives = 87/157 (55%), Gaps = 16/157 (10%)
Query 7 GTDVDD-----DDEDSWCCCEPLN------STERILGWLTCFLGGLILSAVSMGAFNDML 55
G +V+D D+E S E LN + +R G+ C GL + +SM F + +
Sbjct 16 GMEVEDEQQAADEESSLSFMEDLNRNCALTTKQRFYGFAICLSAGLTCTLLSMLVFFNPV 75
Query 56 LGKNNKFAVTYTIGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSSVY 115
KF +T+T+GN++ L T+FL+GP +Q M D +R+ + +++ S++ L ++Y
Sbjct 76 -----KFGITFTLGNLMALGSTAFLIGPQRQVTMMLDPARIYATALYLASIIIALFCALY 130
Query 116 LKVGLVIVFFVCIQWLAYMWYSLSYIPYGQRAVLWIF 152
++ L+ + + +++ +WYSLSYIP+ + V IF
Sbjct 131 VRNKLLTLLAIILEFTGLIWYSLSYIPFARTMVSKIF 167
> 7296331
Length=163
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 55/142 (38%), Positives = 83/142 (58%), Gaps = 12/142 (8%)
Query 8 TDVDDDDEDSWCCCEPLNSTERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYT 67
T ++D SW + RI G++ CF G++LS + A L FAV YT
Sbjct 23 TQINDMSTLSW--------STRIKGFVICFALGILLSLLGSVAL--FLHRGIVVFAVFYT 72
Query 68 IGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSS-VYLKVGLVIVFFV 126
+GN+I +A T FL+GP +Q + M ++RL+ + I + +V T +++ V+ K GL ++F +
Sbjct 73 LGNVISMASTCFLMGPFKQIKKMFAETRLIATIIVLVMMVLTFIAAIVWKKAGLTLIF-I 131
Query 127 CIQWLAYMWYSLSYIPYGQRAV 148
IQ LA WYSLSYIPY + AV
Sbjct 132 IIQSLAMTWYSLSYIPYARDAV 153
> Hs21553317
Length=159
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 80/143 (55%), Gaps = 7/143 (4%)
Query 12 DDDEDSWCCCEPLNSTE-----RILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTY 66
DDE+ + L+++ R+ + CF+ G+ S + G L G FAV Y
Sbjct 11 QDDEEQGLTAQVLDASSLSFNTRLKWFAICFVCGVFFSILGTGLL--WLPGGIKLFAVFY 68
Query 67 TIGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFV 126
T+GN+ LA T FL+GP++Q + M + +RL+ + + + + TL ++++ + V F
Sbjct 69 TLGNLAALASTCFLMGPVKQLKKMFEATRLLATIVMLLCFIFTLCAALWWHKKGLAVLFC 128
Query 127 CIQWLAYMWYSLSYIPYGQRAVL 149
+Q+L+ WYSLSYIPY + AV+
Sbjct 129 ILQFLSMTWYSLSYIPYARDAVI 151
> Hs14736892
Length=160
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 48/148 (32%), Positives = 76/148 (51%), Gaps = 10/148 (6%)
Query 6 DGTDVDDDDEDSWCC-CEPLNSTERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNK--- 61
G D +D S L+ + RI G++ CF G++ S + +LL K
Sbjct 9 SGQDTEDRSGLSEVVEASSLSWSTRIKGFIACFAIGILCSLL-----GTVLLWVPRKGLH 63
Query 62 -FAVTYTIGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSSVYLKVGL 120
FAV YT GNI + T FL+GP++Q + M + +RL+ + + + TL S+ +
Sbjct 64 LFAVFYTFGNIASIGSTIFLMGPVKQLKRMFEPTRLIATIMVLLCFALTLCSAFWWHNKG 123
Query 121 VIVFFVCIQWLAYMWYSLSYIPYGQRAV 148
+ + F +Q LA WYSLS+IP+ + AV
Sbjct 124 LALIFCILQSLALTWYSLSFIPFARDAV 151
> At5g24170
Length=138
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 66/124 (53%), Gaps = 8/124 (6%)
Query 12 DDDEDSWCCCEPLNSTERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTIGNI 71
+D + C L++T+R+ G+ GL+L +SM F G KFA+ +T GN+
Sbjct 23 EDGSEGLC---ALSTTQRMYGFAASLATGLLLMFLSMIVF-----GIPIKFALLFTFGNV 74
Query 72 IGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFVCIQWL 131
+ + T+FL+GP QQ M D R + + I++G +V L+ ++ + ++ V + +
Sbjct 75 LAIGSTAFLMGPEQQMSMMFDPVRFLATSIYIGCVVVALICALLIHSKILTVLAILCEIC 134
Query 132 AYMW 135
A +W
Sbjct 135 ALIW 138
> SPAC1527.02
Length=201
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/121 (25%), Positives = 69/121 (57%), Gaps = 1/121 (0%)
Query 28 ERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTIGNIIGLAGTSFLVGPIQQF 87
ER + + C LG L A++ F ++L K KF + +T+G+++ + G + + G + F
Sbjct 66 ERYMLFGICLLGSLACYAIACFMFPVLVL-KPRKFVLLWTMGSLLAVLGFAIVQGFVAHF 124
Query 88 RNMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFVCIQWLAYMWYSLSYIPYGQRA 147
R + RL + + +L+AT+++++ +K ++ + F + L+++ Y +++ P+G R
Sbjct 125 RQLTTMERLPITLSYFVTLLATIIATIKIKSTILSIVFGVLHILSFVAYLIAFFPFGTRT 184
Query 148 V 148
V
Sbjct 185 V 185
> YBL102w
Length=215
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 69/136 (50%), Gaps = 4/136 (2%)
Query 9 DVDDDDEDSWCCCEPLNSTERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTI 68
D+ D E SW L+ TER++ ++ CFL G +L K KF + +T+
Sbjct 63 DLVQDQEPSWF---QLSRTERMVLFV-CFLLGATACFTLCTFLFPVLAAKPRKFGLLWTM 118
Query 69 GNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFVCI 128
G+++ + L+GP+ +++ + RL S F + T+ + + K ++ + +
Sbjct 119 GSLLFVLAFGVLMGPLAYLKHLTARERLPFSMFFFATCFMTIYFAAFSKNTVLTITCALL 178
Query 129 QWLAYMWYSLSYIPYG 144
+ +A ++Y++SY P+G
Sbjct 179 ELVAVIYYAISYFPFG 194
> CE20520
Length=235
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 67/128 (52%), Gaps = 1/128 (0%)
Query 24 LNSTERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTIGNIIGLAGTSFLVGP 83
+ T+RI+ + C +G I + ++L KFA T+G+++ L +FL+GP
Sbjct 90 MTRTQRIIAFFMCIIGA-IFCFSTAAVLIPVILVSTRKFAGLNTLGSLLLLLSFAFLLGP 148
Query 84 IQQFRNMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFVCIQWLAYMWYSLSYIPY 143
+MA R + + ++ +L ATL SS++LK + + Q +WY LSY+P
Sbjct 149 KSYLTHMASPQRRLVTVSYLSALFATLYSSLWLKSTIFTLIAAIFQGFTLVWYVLSYVPG 208
Query 144 GQRAVLWI 151
G+R + ++
Sbjct 209 GERGLFFM 216
> CE04496
Length=299
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 21/91 (23%), Positives = 41/91 (45%), Gaps = 11/91 (12%)
Query 21 CEPLNSTERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTIGNIIGLAGTSFL 80
+ L + ER +G L L + VS D ++ ++ N A +
Sbjct 204 ADELKNLERQMGELAQMFHDLHIMVVSQAKMVD---------SIVNSVENATEYAKQAR- 253
Query 81 VGPIQQFRNMADKSRLVTSCIFVGSLVATLL 111
G +++ RN+ ++R + CI +GS++A L+
Sbjct 254 -GNVEEARNLQKRARKMKVCIIIGSIIAVLI 283
> ECU09g0940
Length=171
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 42/87 (48%), Gaps = 0/87 (0%)
Query 62 FAVTYTIGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSSVYLKVGLV 121
F + YTI N + FL+G + K + V S F+G + TL + L+
Sbjct 75 FILPYTISNFLFFIMFGFLLGFRSYLEGLFSKKKRVHSSWFIGCTLLTLYVVLKYDRYLL 134
Query 122 IVFFVCIQWLAYMWYSLSYIPYGQRAV 148
+ F IQ ++++ +SL++IP G V
Sbjct 135 NLAFCFIQVVSFIMFSLTFIPGGTSGV 161
> At2g39480
Length=1407
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 34/86 (39%), Gaps = 15/86 (17%)
Query 33 WLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTIGNIIGLAGTSFLVGPIQQFRNMAD 92
WL LG + A G+FN +L +I L T++ R D
Sbjct 832 WLYAVLGSI--GAAIFGSFNPLL-------------AYVIALVVTTYYTSKGSHLREEVD 876
Query 93 KSRLVTSCIFVGSLVATLLSSVYLKV 118
K L+ +C+ + ++VA L Y +
Sbjct 877 KWCLIIACMGIVTVVANFLQHFYFGI 902
> CE27888
Length=550
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 56/114 (49%), Gaps = 10/114 (8%)
Query 33 WLTCFLGGLILSAVSMGAFNDMLLGKNNKF----AVTYTIGNIIGLAGTSFLVGPIQQFR 88
+L C ++LS + N ++ +++K A +Y IGNII G+ + P R
Sbjct 7 FLNCLFPPVVLSEPKIIPENTQIMDESHKMGFLGATSYVIGNII---GSGIFITPASILR 63
Query 89 NMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFVC-IQWLAYM-WYSLSY 140
N+ D L + +++A L + Y+++G I C ++ Y+ WYS+++
Sbjct 64 NV-DSIGLSLLIWVLCAVIAILGAICYIELGTSIREAGCDFAYICYVKWYSIAF 116
> CE09028
Length=566
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 56/114 (49%), Gaps = 10/114 (8%)
Query 33 WLTCFLGGLILSAVSMGAFNDMLLGKNNKF----AVTYTIGNIIGLAGTSFLVGPIQQFR 88
+L C ++LS + N ++ +++K A +Y IGNII G+ + P R
Sbjct 7 FLNCLFPPVVLSEPKIIPENTQIMDESHKMGFLGATSYVIGNII---GSGIFITPASILR 63
Query 89 NMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFVC-IQWLAYM-WYSLSY 140
N+ D L + +++A L + Y+++G I C ++ Y+ WYS+++
Sbjct 64 NV-DSIGLSLLIWVLCAVIAILGAICYIELGTSIREAGCDFAYICYVKWYSIAF 116
> Hs7705636
Length=138
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/46 (28%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 67 TIGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLS 112
IGN++ +AG +F++G + FR K ++ + F+G + L+
Sbjct 36 AIGNVLFVAGLAFVIGLERTFRFFFQKHKMKATGFFLGGVFVVLIG 81
> CE10200
Length=310
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 36/136 (26%), Positives = 53/136 (38%), Gaps = 21/136 (15%)
Query 2 PLFADGTDVDDDDEDSWCCCEPLNSTERILGWLTCFLGGLILSAVSMGAFNDMLLGK--- 58
P D D E+ + C + L +TC + GL L A+S N ++ K
Sbjct 151 PFLFDHVPYPDYQENDFKCTDTL---------ITCLVSGLFLLAISTNFMNILIAVKLFC 201
Query 59 ---NNKFAVTYTIGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSS-- 113
+ + T N + FL Q + M D + TS F GS V +L S
Sbjct 202 LSKSTSSLSSETAKNRRKMKIRFFLQSCFQDWICMLDVAMNFTSIEFCGSHVCAVLISMG 261
Query 114 ----VYLKVGLVIVFF 125
VY+ GL++ FF
Sbjct 262 FDVLVYVMDGLIMYFF 277
> At5g22930
Length=248
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 74 LAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLL 111
++GT ++ P+ Q + + +K +L+ + I G L+ TLL
Sbjct 152 VSGTEIIMDPLLQDKPLPEKGKLLQAVIKAGPLLQTLL 189
> At3g55320
Length=1408
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 33/86 (38%), Gaps = 15/86 (17%)
Query 33 WLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTIGNIIGLAGTSFLVGPIQQFRNMAD 92
WL LG L A G+FN +L +I L T + R D
Sbjct 833 WLYAVLGSL--GAAIFGSFNPLL-------------AYVIALVVTEYYKSKGGHLREEVD 877
Query 93 KSRLVTSCIFVGSLVATLLSSVYLKV 118
K L+ +C+ + ++VA L Y +
Sbjct 878 KWCLIIACMGIVTVVANFLQHFYFGI 903
> SPAC16E8.07c
Length=805
Score = 28.5 bits (62), Expect = 6.4, Method: Composition-based stats.
Identities = 10/17 (58%), Positives = 13/17 (76%), Gaps = 0/17 (0%)
Query 115 YLKVGLVIVFFVCIQWL 131
YL+VGLVI +C+ WL
Sbjct 620 YLQVGLVIAALICVPWL 636
> CE27805
Length=791
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 32/145 (22%), Positives = 57/145 (39%), Gaps = 29/145 (20%)
Query 8 TDVDDDDEDSWCCCEPLN-STERILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTY 66
T V+++DE +P +T GW I+ V+ F+ +L+ N+ T
Sbjct 144 TYVNENDEACQVVSDPGKIATHYFKGWF-------IIDMVAAVPFDLLLVSTNSDETTT- 195
Query 67 TIGNIIGLAGTSFLVGPIQQFRNMADKSRLVTSCIFVGSLVATLLSSVYLKVGLVIVFFV 126
+IGL T+ L+ ++ R + S G+ V LL + F +
Sbjct 196 ----LIGLLKTARLLRLVRVARKLDRYSEY-------GAAVLLLLMAT---------FAL 235
Query 127 CIQWLAYMWYSLSYIPYGQRAVLWI 151
WLA +WY++ + W+
Sbjct 236 IAHWLACIWYAIGSAELSHKEYTWL 260
> CE28039
Length=466
Score = 27.7 bits (60), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 7/43 (16%)
Query 30 ILGWLTCFLGGLILSAVSMGAFNDMLLGKNNKFAVTYTIGNII 72
++ W++ +LG + A +M LG N+ A+T+ GNII
Sbjct 255 VISWISFYLGPRAIPARTM-------LGVNSLLAMTFQFGNII 290
Lambda K H
0.328 0.143 0.465
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2100092188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40