bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4181_orf2
Length=212
Score E
Sequences producing significant alignments: (Bits) Value
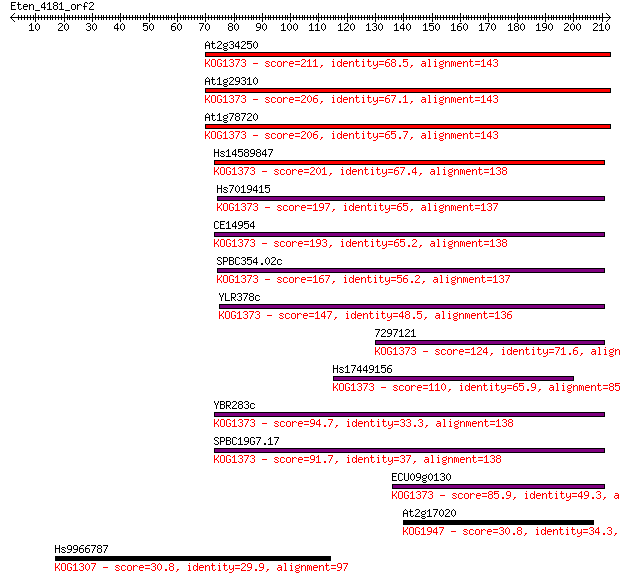
At2g34250 211 6e-55
At1g29310 206 2e-53
At1g78720 206 2e-53
Hs14589847 201 7e-52
Hs7019415 197 1e-50
CE14954 193 2e-49
SPBC354.02c 167 1e-41
YLR378c 147 2e-35
7297121 124 1e-28
Hs17449156 110 2e-24
YBR283c 94.7 1e-19
SPBC19G7.17 91.7 1e-18
ECU09g0130 85.9 5e-17
At2g17020 30.8 1.9
Hs9966787 30.8 2.3
> At2g34250
Length=475
Score = 211 bits (538), Expect = 6e-55, Method: Compositional matrix adjust.
Identities = 98/143 (68%), Positives = 120/143 (83%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
M G R L LV+P + LPEV + +R+V FREKV++T+++L +FLVC Q+PL+GI G
Sbjct 1 MGGGFRVLHLVRPFLAFLPEVQSADRKVPFREKVIYTVISLFIFLVCSQLPLYGIHSTTG 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYWMRVILASNRGT+MELGI+PIVTSG+VMQLLAGS+II+VD ++REDRAL GAQK
Sbjct 61 ADPFYWMRVILASNRGTVMELGITPIVTSGLVMQLLAGSKIIEVDNNVREDRALLNGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL +LI I EAVAYV+SGMYGPV
Sbjct 121 LLGILIAIGEAVAYVLSGMYGPV 143
> At1g29310
Length=475
Score = 206 bits (525), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 96/143 (67%), Positives = 119/143 (83%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
M G R L LV+P + LPEV + +R++ FREKV++T+++L +FLVC Q+PL+GI G
Sbjct 1 MGGGFRVLHLVRPFLAFLPEVQSADRKIPFREKVIYTVISLFIFLVCSQLPLYGIHSTTG 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYWMRVILASNRGT+MELGI+PIVTSG+VMQLLAGS+II+VD ++REDRAL GAQK
Sbjct 61 ADPFYWMRVILASNRGTVMELGITPIVTSGLVMQLLAGSKIIEVDNNVREDRALLNGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL +L I EAVAYV+SGMYGPV
Sbjct 121 LLWILSAIGEAVAYVLSGMYGPV 143
> At1g78720
Length=475
Score = 206 bits (525), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 94/143 (65%), Positives = 120/143 (83%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
M G R + LV+P + LPEV +PER++ FREKV++T+++L +FLVC Q+PL+GI G
Sbjct 1 MVGGFRVIHLVRPFLAFLPEVQSPERKIPFREKVIYTVISLFIFLVCSQLPLYGIHSTTG 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYWMRVILAS+RGT+MELGI+PIVTSGMVMQLLAGS+II++D ++REDRAL GAQK
Sbjct 61 ADPFYWMRVILASSRGTVMELGITPIVTSGMVMQLLAGSKIIEIDNNVREDRALLNGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL +LI + +AVAYV+SGMYG V
Sbjct 121 LLGILIAVGQAVAYVLSGMYGSV 143
> Hs14589847
Length=476
Score = 201 bits (512), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 93/138 (67%), Positives = 114/138 (82%), Gaps = 1/138 (0%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADP 132
GI+FL ++KP VLPE+ PER++ FREKVLWT + L +FLVCCQIPLFGI ADP
Sbjct 2 GIKFLEVIKPFCAVLPEIQKPERKIQFREKVLWTAITLFIFLVCCQIPLFGIMSSDSADP 61
Query 133 FYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLA 192
FYWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + +DRALF GAQKL
Sbjct 62 FYWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDT-PKDRALFNGAQKLFG 120
Query 193 LLITIXEAVAYVISGMYG 210
++ITI +A+ YV++GMYG
Sbjct 121 MIITIGQAIVYVMTGMYG 138
> Hs7019415
Length=476
Score = 197 bits (502), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 113/137 (82%), Gaps = 1/137 (0%)
Query 74 IRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPF 133
I+FL ++KP +LPE+ PER++ F+EKVLWT + L +FLVCCQIPLFGI ADPF
Sbjct 3 IKFLEVIKPFCVILPEIQKPERKIQFKEKVLWTAITLFIFLVCCQIPLFGIMSSDSADPF 62
Query 134 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLAL 193
YWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL +
Sbjct 63 YWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFGM 121
Query 194 LITIXEAVAYVISGMYG 210
+ITI +++ YV++GMYG
Sbjct 122 IITIGQSIVYVMTGMYG 138
> CE14954
Length=473
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 90/138 (65%), Positives = 112/138 (81%), Gaps = 1/138 (0%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADP 132
GI+FL VKP +PEV PER++ FREK+LWT + L VFLVCCQIPLFGI ADP
Sbjct 2 GIKFLEFVKPFCGFVPEVSKPERKIQFREKMLWTAITLFVFLVCCQIPLFGIMSTDSADP 61
Query 133 FYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLA 192
FYW+RVI+ASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL
Sbjct 62 FYWLRVIMASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFG 120
Query 193 LLITIXEAVAYVISGMYG 210
++IT+ +A+ YV+SG+YG
Sbjct 121 MVITVGQAIVYVMSGLYG 138
> SPBC354.02c
Length=479
Score = 167 bits (423), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 77/137 (56%), Positives = 105/137 (76%), Gaps = 0/137 (0%)
Query 74 IRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPF 133
+RFL LVKP LPE+ APER+V F++K+LWT + L +FLV Q+PL+GI +DP
Sbjct 4 LRFLDLVKPFAPFLPEIAAPERKVPFKQKMLWTGVTLLIFLVMSQVPLYGIVSSDSSDPL 63
Query 134 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLAL 193
W+R+ILA+NRGTLMELGISPIVTS M++QLL GS++I+V+ L+ DR ++Q QK LA+
Sbjct 64 LWLRMILAANRGTLMELGISPIVTSSMLVQLLVGSQLIEVNMELKSDREMYQLVQKFLAI 123
Query 194 LITIXEAVAYVISGMYG 210
+I +A AYV++GMYG
Sbjct 124 IIAFGQATAYVLTGMYG 140
> YLR378c
Length=480
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 66/136 (48%), Positives = 99/136 (72%), Gaps = 0/136 (0%)
Query 75 RFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPFY 134
R L L KP LPEV APER+V + +K++WT ++L +FL+ QIPL+GI +DP Y
Sbjct 5 RVLDLFKPFESFLPEVIAPERKVPYNQKLIWTGVSLLIFLILGQIPLYGIVSSETSDPLY 64
Query 135 WMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLALL 194
W+R +LASNRGTL+ELG+SPI+TS M+ Q L G++++Q+ ++DR LFQ AQK+ A++
Sbjct 65 WLRAMLASNRGTLLELGVSPIITSSMIFQFLQGTQLLQIRPESKQDRELFQIAQKVCAII 124
Query 195 ITIXEAVAYVISGMYG 210
+ + +A+ V++G YG
Sbjct 125 LILGQALVVVMTGNYG 140
> 7297121
Length=423
Score = 124 bits (311), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 58/81 (71%), Positives = 72/81 (88%), Gaps = 1/81 (1%)
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYW+RVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + +DRALF GAQK
Sbjct 6 ADPFYWIRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDT-PKDRALFNGAQK 64
Query 190 LLALLITIXEAVAYVISGMYG 210
L ++ITI +A+ YV++GMYG
Sbjct 65 LFGMVITIGQAIVYVMTGMYG 85
> Hs17449156
Length=114
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 56/87 (64%), Positives = 68/87 (78%), Gaps = 3/87 (3%)
Query 115 VCCQIPLFGIRGGRGADPFYWMRVILASNRGTLM--ELGISPIVTSGMVMQLLAGSRIIQ 172
+CCQIPLFGI DPFYWMRVILASNRGTLM ELGISPIV SG++MQLLAG++II+
Sbjct 25 MCCQIPLFGIMSSDSVDPFYWMRVILASNRGTLMELELGISPIVMSGLIMQLLAGAKIIE 84
Query 173 VDQSLREDRALFQGAQKLLALLITIXE 199
V + +D+ L AQKLL ++ITI +
Sbjct 85 VGGT-PKDQVLINEAQKLLHMIITIGQ 110
> YBR283c
Length=490
Score = 94.7 bits (234), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/141 (32%), Positives = 84/141 (59%), Gaps = 4/141 (2%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG--- 129
G R + +VKP + +LPEV P ++ F +K+++T+ A ++L Q PL G+
Sbjct 3 GFRLIDIVKPILPILPEVELPFEKLPFDDKIVYTIFAGLIYLFA-QFPLVGLPKATTPNV 61
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
DP Y++R + TL+E G+ P ++SG+++QLLAG ++I+V+ ++ DR LFQ K
Sbjct 62 NDPIYFLRGVFGCEPRTLLEFGLFPNISSGLILQLLAGLKVIKVNFKIQSDRELFQSLTK 121
Query 190 LLALLITIXEAVAYVISGMYG 210
+ A++ + ++ +G +G
Sbjct 122 VFAIVQYVILTNIFIFAGYFG 142
> SPBC19G7.17
Length=179
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 51/138 (36%), Positives = 78/138 (56%), Gaps = 0/138 (0%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADP 132
G RF++ +KP +LPEV P+ + EK+ W + V+ + IP++G DP
Sbjct 3 GARFINFIKPLSSLLPEVEGPKTHLELVEKLGWMAGCVVVYQILSIIPVYGAEKTDTLDP 62
Query 133 FYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLA 192
RV+ S+ LM G++PI S ++Q+LA + I V+ +L DR LFQ AQK+++
Sbjct 63 INNFRVLDGSSASGLMITGLAPIYLSSFLLQILASKKKIAVNFNLIIDRVLFQNAQKVVS 122
Query 193 LLITIXEAVAYVISGMYG 210
L+ + AV YV SG YG
Sbjct 123 ALLYLILAVTYVSSGYYG 140
> ECU09g0130
Length=410
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 37/75 (49%), Positives = 57/75 (76%), Gaps = 0/75 (0%)
Query 136 MRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLALLI 195
MR ++ASNRGTLM+LG SP+VTS ++MQ L S I++VD S++ED+ L Q+L++L++
Sbjct 1 MRAMMASNRGTLMDLGTSPVVTSSLIMQFLTMSEILKVDWSIKEDKNLHNATQRLISLIM 60
Query 196 TIXEAVAYVISGMYG 210
T+ +A+ V +G YG
Sbjct 61 TVGQALVQVYTGFYG 75
> At2g17020
Length=656
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 33/67 (49%), Gaps = 3/67 (4%)
Query 140 LASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLALLITIXE 199
L S+R T +ELG +TS M+ QLL + I D + + Q Q+L + I +
Sbjct 192 LVSDRLTHLELGH---ITSRMMTQLLTSTEISGQDSNRVTTSTVLQNVQRLRLSVDCITD 248
Query 200 AVAYVIS 206
AV IS
Sbjct 249 AVVKAIS 255
> Hs9966787
Length=661
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 39/100 (39%), Gaps = 14/100 (14%)
Query 17 APSSQAPLWRRAAAQRSSSSSSSGAAQQQLAGRSACAPPSSLPFFLSLASPRKMAKGIRF 76
PSS A Q + S + A+ Q A P LSLA P + K + F
Sbjct 425 TPSSDA----SEPVQNGNLSHNIEGAEAQTADEEEDQP-------LSLAWPSETRKQVTF 473
Query 77 L---SLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVF 113
L +V P LP+V P R F ++ +AVF
Sbjct 474 LIVFPIVFPLWITLPDVRKPSSRKFFPITFFGSITWIAVF 513
Lambda K H
0.325 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3861735048
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40