bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4144_orf3
Length=142
Score E
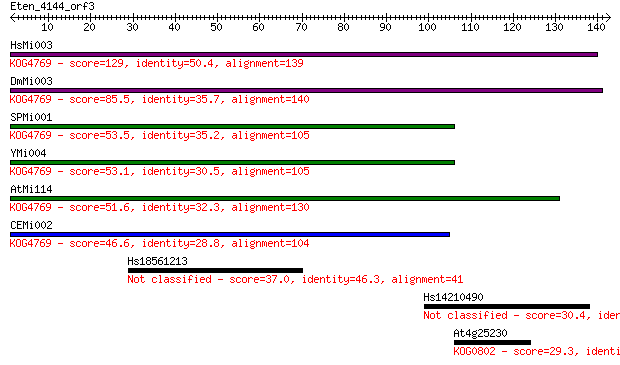
Sequences producing significant alignments: (Bits) Value
HsMi003 129 2e-30
DmMi003 85.5 3e-17
SPMi001 53.5 1e-07
YMi004 53.1 1e-07
AtMi114 51.6 5e-07
CEMi002 46.6 2e-05
Hs18561213 37.0 0.011
Hs14210490 30.4 1.1
At4g25230 29.3 2.8
> HsMi003
Length=513
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 70/139 (50%), Positives = 80/139 (57%), Gaps = 0/139 (0%)
Query 1 VLANFPYFFPKGGVFFFLGGFFHCFPLFSGFFFKDTGAKIPFPIFFGGANKTFFPQPFLG 60
V+A+F Y G VF +GGF H FPLFSG+ T AKI F I F G N TFFPQ FLG
Sbjct 373 VVAHFHYVLSMGAVFAIMGGFIHWFPLFSGYTLDQTYAKIHFTIMFIGVNLTFFPQHFLG 432
Query 61 FLGNPGGFSASPGASPIGNFFSSKGSFFSLTAVFLMFFFFGEAFAFKGKFLPLDLPPQIL 120
G P +S P A N SS GSF SLTAV LM F EAFA K K L ++ P L
Sbjct 433 LSGMPRRYSDYPDAYTTWNILSSVGSFISLTAVMLMIFMIWEAFASKRKVLMVEEPSMNL 492
Query 121 EGLKGAPPPFPHLEDPPFF 139
E L G PPP+ E+P +
Sbjct 493 EWLYGCPPPYHTFEEPVYM 511
> DmMi003
Length=511
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 50/141 (35%), Positives = 66/141 (46%), Gaps = 2/141 (1%)
Query 1 VLANFPYFFPKGGVFFFLGGFFHCFPLFSGFFFKDTGAKIPFPIFFGGANKTFFPQPFLG 60
V+A+F Y G VF + GF H +PLF+G + K F I F G N TFFPQ FLG
Sbjct 371 VVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLG 430
Query 61 FLGNPGGFSASPGASPIGNFFSSKGSFFSLTAVFLMFFFFGEAFAFKGKFL-PLDLPPQI 119
G P +S P A N S+ GS SL + FF E+ + + + P+ L I
Sbjct 431 LAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSI 490
Query 120 LEGLKGAPPPFPHLEDPPFFT 140
E + PP + P T
Sbjct 491 -EWYQNTPPAEHSYSELPLLT 510
> SPMi001
Length=537
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/105 (35%), Positives = 50/105 (47%), Gaps = 1/105 (0%)
Query 1 VLANFPYFFPKGGVFFFLGGFFHCFPLFSGFFFKDTGAKIPFPIFFGGANKTFFPQPFLG 60
V+A+F Y G +F G ++ +F G + +T A I F I F G N F PQ FLG
Sbjct 380 VVAHFHYVLSMGALFGLCGAYYWSPKMF-GLMYNETLASIQFWILFIGVNIVFGPQHFLG 438
Query 61 FLGNPGGFSASPGASPIGNFFSSKGSFFSLTAVFLMFFFFGEAFA 105
G P P A NF SS GS S+ ++FL + + F
Sbjct 439 LNGMPRRIPDYPEAFVGWNFVSSIGSVISILSLFLFMYVMYDQFT 483
> YMi004
Length=534
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 49/105 (46%), Gaps = 0/105 (0%)
Query 1 VLANFPYFFPKGGVFFFLGGFFHCFPLFSGFFFKDTGAKIPFPIFFGGANKTFFPQPFLG 60
V+ +F Y G +F G+++ P G + + A+I F + F GAN FFP FLG
Sbjct 373 VVGHFHYVLSMGAIFSLFAGYYYWSPQILGLNYNEKLAQIQFWLIFIGANVIFFPMHFLG 432
Query 61 FLGNPGGFSASPGASPIGNFFSSKGSFFSLTAVFLMFFFFGEAFA 105
G P P A N+ +S GSF + ++FL + +
Sbjct 433 INGMPRRIPDYPDAFAGWNYVASIGSFIATLSLFLFIYILYDQLV 477
> AtMi114
Length=527
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 56/136 (41%), Gaps = 6/136 (4%)
Query 1 VLANFPYFFPKGGVFFFLGGFFHCFPLFSGFFFKDTGAKIPFPIFFGGANKTFFPQPFLG 60
V+A+F Y G VF GF++ G + +T +I F I F G N TFFP FLG
Sbjct 375 VVAHFHYVLSMGAVFALFAGFYYWVGKIFGRTYPETLGQIHFWITFFGVNLTFFPMHFLG 434
Query 61 FLGNPGGFSASPGASPIGNFFSSKGSFFSLTAVFLMFFFFGEAFAFKGKF------LPLD 114
G P P A N SS GS+ S+ + F + L+
Sbjct 435 LSGMPRRIPDYPDAYAGWNALSSFGSYISVVGICCFFVVVTITLSSGNNKRCAPSPWALE 494
Query 115 LPPQILEGLKGAPPPF 130
L LE + +PP F
Sbjct 495 LNSTTLEWMVQSPPAF 510
> CEMi002
Length=525
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 43/104 (41%), Gaps = 0/104 (0%)
Query 1 VLANFPYFFPKGGVFFFLGGFFHCFPLFSGFFFKDTGAKIPFPIFFGGANKTFFPQPFLG 60
V+++F Y G VF G + +G+ F + F G N TFFP F G
Sbjct 380 VVSHFHYVLSLGAVFGIFTGVTLWWSFITGYVLDKLMMSAVFILLFIGVNLTFFPLHFAG 439
Query 61 FLGNPGGFSASPGASPIGNFFSSKGSFFSLTAVFLMFFFFGEAF 104
G P + P + N +S GS S +FL + E+F
Sbjct 440 LHGFPRKYLDYPDVYSVWNIIASYGSIISTAGLFLFIYVLLESF 483
> Hs18561213
Length=118
Score = 37.0 bits (84), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 29 SGFFFKDTGAKIPFPIFFGGANKTFFPQPFLGFLGNPGGFS 69
G+ AK F I F G N TFFPQ FLG G P +S
Sbjct 39 QGYILNQAYAKAHFAIIFIGVNLTFFPQHFLGLSGMPQRYS 79
> Hs14210490
Length=530
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 99 FFGEAFAFKGKFLPLDLPPQILEGLKGAPPPFPHLEDPP 137
FF A+A G L L+L P+ GL PP P +E P
Sbjct 287 FFSPAYAASGGVLGLNLQPEQPHGLYLLPPGAPFIELLP 325
> At4g25230
Length=594
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 10/18 (55%), Positives = 14/18 (77%), Gaps = 0/18 (0%)
Query 106 FKGKFLPLDLPPQILEGL 123
+KG FLPL +PP I +G+
Sbjct 104 YKGTFLPLVIPPTIFQGV 121
Lambda K H
0.333 0.156 0.527
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40