bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4079_orf1
Length=85
Score E
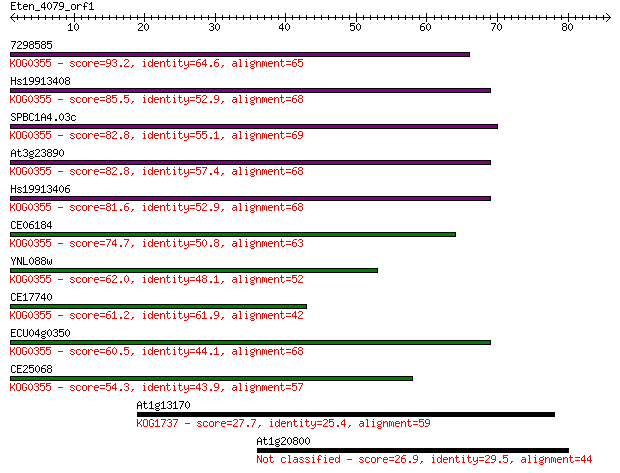
Sequences producing significant alignments: (Bits) Value
7298585 93.2 1e-19
Hs19913408 85.5 2e-17
SPBC1A4.03c 82.8 1e-16
At3g23890 82.8 2e-16
Hs19913406 81.6 3e-16
CE06184 74.7 3e-14
YNL088w 62.0 3e-10
CE17740 61.2 4e-10
ECU04g0350 60.5 8e-10
CE25068 54.3 6e-08
At1g13170 27.7 5.4
At1g20800 26.9 9.8
> 7298585
Length=1423
Score = 93.2 bits (230), Expect = 1e-19, Method: Composition-based stats.
Identities = 42/65 (64%), Positives = 49/65 (75%), Gaps = 0/65 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAKSPILEGVLLWA 60
+R HLWVFV CLIENP FDSQTKE +T FGSKC LSE+ IN ++KS I+E VL WA
Sbjct 313 VRNHLWVFVNCLIENPTFDSQTKENMTLQQKGFGSKCTLSEKFINNMSKSGIVESVLAWA 372
Query 61 QTKAQ 65
+ KAQ
Sbjct 373 KFKAQ 377
> Hs19913408
Length=1621
Score = 85.5 bits (210), Expect = 2e-17, Method: Composition-based stats.
Identities = 36/68 (52%), Positives = 46/68 (67%), Gaps = 0/68 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAKSPILEGVLLWA 60
++ H+WVF+ CLIENP FDSQTKE +T FGSKC LSE+ A + I+E +L W
Sbjct 372 VKNHIWVFINCLIENPTFDSQTKENMTLQPKSFGSKCQLSEKFFKAASNCGIVESILNWV 431
Query 61 QTKAQVQL 68
+ KAQ QL
Sbjct 432 KFKAQTQL 439
> SPBC1A4.03c
Length=1485
Score = 82.8 bits (203), Expect = 1e-16, Method: Composition-based stats.
Identities = 38/69 (55%), Positives = 52/69 (75%), Gaps = 0/69 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAKSPILEGVLLWA 60
++ ++ VFV C IENP+FDSQTKETLTT VS FGS+C LS++ + A+ KS ++E VL +A
Sbjct 401 IKNYVQVFVNCQIENPSFDSQTKETLTTKVSAFGSQCTLSDKFLKAIKKSSVVEEVLKFA 460
Query 61 QTKAQVQLT 69
KA QL+
Sbjct 461 TAKADQQLS 469
> At3g23890
Length=1473
Score = 82.8 bits (203), Expect = 2e-16, Method: Composition-based stats.
Identities = 39/68 (57%), Positives = 46/68 (67%), Gaps = 0/68 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAKSPILEGVLLWA 60
++ HLWVFV LI+NPAFDSQTKETLT S FGSKC LSE + V KS ++E +L WA
Sbjct 358 VKNHLWVFVNALIDNPAFDSQTKETLTLRQSSFGSKCELSEDFLKKVGKSGVVENLLSWA 417
Query 61 QTKAQVQL 68
K L
Sbjct 418 DFKQNKDL 425
> Hs19913406
Length=1531
Score = 81.6 bits (200), Expect = 3e-16, Method: Composition-based stats.
Identities = 36/68 (52%), Positives = 45/68 (66%), Gaps = 0/68 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAKSPILEGVLLWA 60
++ H+W+FV LIENP FDSQTKE +T FGS C LSE+ I A I+E +L W
Sbjct 356 VKNHMWIFVNALIENPTFDSQTKENMTLQPKSFGSTCQLSEKFIKAAIGCGIVESILNWV 415
Query 61 QTKAQVQL 68
+ KAQVQL
Sbjct 416 KFKAQVQL 423
> CE06184
Length=1520
Score = 74.7 bits (182), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/63 (50%), Positives = 42/63 (66%), Gaps = 0/63 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAKSPILEGVLLWA 60
++ H+WVFV CLIENP FDSQTKET+T +FGS C LSE+ A + I + V+ W
Sbjct 391 IKNHMWVFVNCLIENPTFDSQTKETMTLQQKQFGSTCVLSEKFSKAASSVGITDAVMSWV 450
Query 61 QTK 63
+ K
Sbjct 451 RFK 453
> YNL088w
Length=1428
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 25/52 (48%), Positives = 37/52 (71%), Gaps = 0/52 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAKSPI 52
++ ++++F+ CLIENPAF SQTKE LTT V FGS+C + IN + K+ +
Sbjct 345 IKNNMFIFINCLIENPAFTSQTKEQLTTRVKDFGSRCEIPLEYINKIMKTDL 396
> CE17740
Length=654
Score = 61.2 bits (147), Expect = 4e-10, Method: Composition-based stats.
Identities = 26/42 (61%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSER 42
+ HLWVFV CLIE P FDSQTKE +T RFGS C LS++
Sbjct 386 IENHLWVFVNCLIEWPTFDSQTKEMMTLRHDRFGSTCALSKK 427
> ECU04g0350
Length=1144
Score = 60.5 bits (145), Expect = 8e-10, Method: Composition-based stats.
Identities = 30/69 (43%), Positives = 44/69 (63%), Gaps = 1/69 (1%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGSKCNLSERTINAVAK-SPILEGVLLW 59
+R ++VF+ LI+NPAFDSQTKE LT S FGSKC + ++ K +PI++ + +
Sbjct 323 VRSCMFVFINSLIDNPAFDSQTKENLTLRASAFGSKCEPMGDFVKSIIKNTPIVQKITSF 382
Query 60 AQTKAQVQL 68
A+ K QL
Sbjct 383 AKAKQSQQL 391
> CE25068
Length=1114
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 25/59 (42%), Positives = 42/59 (71%), Gaps = 3/59 (5%)
Query 1 LRQHLWVFVKCLIENPAFDSQTKETLTTTVSRFGS--KCNLSERTINAVAKSPILEGVL 57
++ +L +F+ CLIENP+F+SQTKETLTT FGS +C+ +++T +S ++E ++
Sbjct 308 IKNNLSIFINCLIENPSFESQTKETLTTKAKNFGSIFECD-AKKTAEWAEQSGLIEDIV 365
> At1g13170
Length=816
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 15/61 (24%), Positives = 27/61 (44%), Gaps = 2/61 (3%)
Query 19 DSQTKETLTTTVSRFGSKCN--LSERTINAVAKSPILEGVLLWAQTKAQVQLTYATRPHA 76
D++T E + + ++ L + T P+ E VLLW + K+ + Y P A
Sbjct 646 DNRTGEKVAILIGKWDEAMYYVLGDPTTKPKGYDPMTEAVLLWERDKSPTKTRYNLSPFA 705
Query 77 V 77
+
Sbjct 706 I 706
> At1g20800
Length=492
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 36 KCNLSERTINAVAKSPILEGVLLWAQTKAQVQLTYATRPHAVLF 79
K N +RT+ + +S ++EG LL+ T + T P +++F
Sbjct 437 KINKRKRTVRSAGRSGVVEGFLLYVVTAQYCRNTGEKLPISMVF 480
Lambda K H
0.325 0.133 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194132014
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40