bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4067_orf4
Length=176
Score E
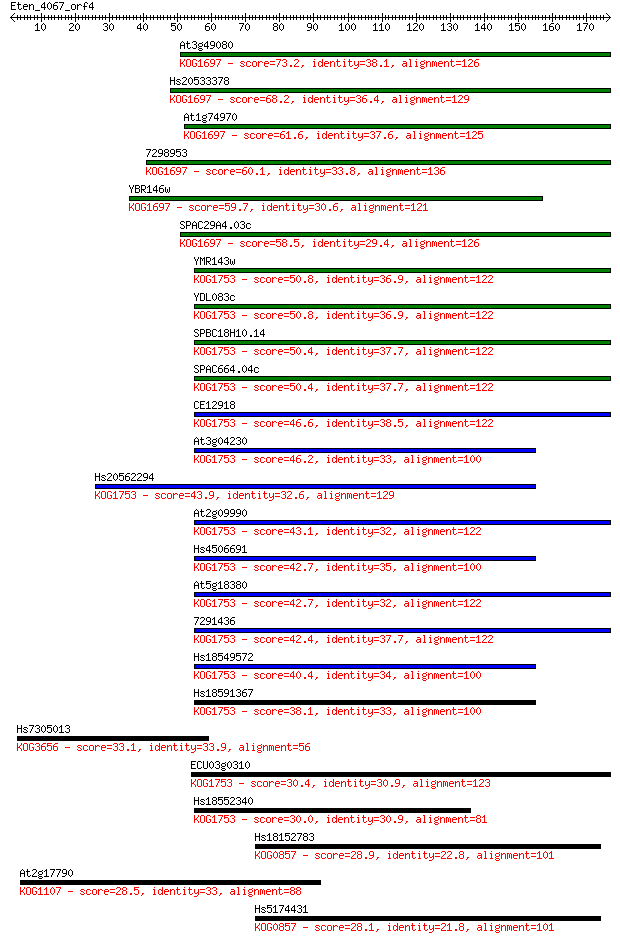
Sequences producing significant alignments: (Bits) Value
At3g49080 73.2 2e-13
Hs20533378 68.2 9e-12
At1g74970 61.6 7e-10
7298953 60.1 2e-09
YBR146w 59.7 3e-09
SPAC29A4.03c 58.5 7e-09
YMR143w 50.8 2e-06
YDL083c 50.8 2e-06
SPBC18H10.14 50.4 2e-06
SPAC664.04c 50.4 2e-06
CE12918 46.6 2e-05
At3g04230 46.2 4e-05
Hs20562294 43.9 1e-04
At2g09990 43.1 3e-04
Hs4506691 42.7 4e-04
At5g18380 42.7 4e-04
7291436 42.4 5e-04
Hs18549572 40.4 0.002
Hs18591367 38.1 0.008
Hs7305013 33.1 0.32
ECU03g0310 30.4 2.0
Hs18552340 30.0 2.2
Hs18152783 28.9 5.3
At2g17790 28.5 7.7
Hs5174431 28.1 9.5
> At3g49080
Length=143
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 48/127 (37%), Positives = 71/127 (55%), Gaps = 3/127 (2%)
Query 51 AMGQGTRKRAAAVALLTRGTGQVRVCGRE-EVYTRWPYVYNRMEVLQPLLLAAAAGLFDV 109
A G G RK + A + G G+ +V +E +VY +P + +R +L+PL G +D+
Sbjct 19 AYGTGRRKCSIARVWIQPGEGKFQVNEKEFDVY--FPMLDHRAALLRPLAETKTLGRWDI 76
Query 110 DIQVRGGGPSGQAGAARLAVARALVAACEDCQPALQEAMMLYEDTRQRMPKMPGRMKARK 169
V+GGG +GQ GA +L ++RAL D + +L+ A L D+R K PG+ KARK
Sbjct 77 KCTVKGGGTTGQVGAIQLGISRALQNWEPDMRTSLRAAGFLTRDSRVVERKKPGKAKARK 136
Query 170 QRQWSKR 176
QW KR
Sbjct 137 SFQWVKR 143
> Hs20533378
Length=396
Score = 68.2 bits (165), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 47/132 (35%), Positives = 75/132 (56%), Gaps = 5/132 (3%)
Query 48 LCTAMGQGTRKRAAAVALLTR-GTGQVRVCGRE-EVYTRWPYVYNRMEVLQPLLLAAAAG 105
+ + +G RK A A A++ + G+G+++V G + ++Y +P +R +++ P G
Sbjct 267 MAFSKSEGKRKTAKAEAIVYKHGSGRIKVNGIDYQLY--FPITQDREQLMFPFHFVDRLG 324
Query 106 LFDVDIQVRGGGPSGQAGAARLAVARALVA-ACEDCQPALQEAMMLYEDTRQRMPKMPGR 164
DV V GGG S QAGA RLA+A+AL + ED +++A +L D R R K PG+
Sbjct 325 KHDVTCTVSGGGRSAQAGAIRLAMAKALCSFVTEDEVEWMRQAGLLTTDPRVRERKKPGQ 384
Query 165 MKARKQRQWSKR 176
AR++ W KR
Sbjct 385 EGARRKFTWKKR 396
> At1g74970
Length=208
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 47/127 (37%), Positives = 65/127 (51%), Gaps = 4/127 (3%)
Query 52 MGQGTRKRAAAVALLTRGTGQVRVCGRE--EVYTRWPYVYNRMEVLQPLLLAAAAGLFDV 109
+G G RK A A +L GTG+V + R+ E P ++V PL+ +D+
Sbjct 84 IGTGRRKCAIARVVLQEGTGKVIINYRDAKEYLQGNPLWLQYVKV--PLVTLGYENSYDI 141
Query 110 DIQVRGGGPSGQAGAARLAVARALVAACEDCQPALQEAMMLYEDTRQRMPKMPGRMKARK 169
++ GGG SGQA A L VARAL+ D + L++ +L D R K G KARK
Sbjct 142 FVKAHGGGLSGQAQAITLGVARALLKVSADHRSPLKKEGLLTRDARVVERKKAGLKKARK 201
Query 170 QRQWSKR 176
Q+SKR
Sbjct 202 APQFSKR 208
> 7298953
Length=395
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 46/138 (33%), Positives = 73/138 (52%), Gaps = 6/138 (4%)
Query 41 EHGGRVWLCTAMGQGTRKRA-AAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLL 99
+ GR ++ T + RK A A V + GTG++ + G++ Y R ++L PL
Sbjct 262 DEEGRQYITTY--ECLRKTARADVTVRLPGTGKISINGKDISYFEDENC--REQLLFPLQ 317
Query 100 LAAAAGLFDVDIQVRGGGPSGQAGAARLAVARALVAACE-DCQPALQEAMMLYEDTRQRM 158
+ G DV+ V GGGPSGQAGA R +A +L + + + +++ A +L D R+R
Sbjct 318 FSELLGKVDVEANVEGGGPSGQAGAIRWGIAMSLRSFVDQEMIESMRLAGLLTRDYRRRE 377
Query 159 PKMPGRMKARKQRQWSKR 176
K G+ AR++ W KR
Sbjct 378 RKKFGQEGARRKYTWKKR 395
> YBR146w
Length=278
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 63/121 (52%), Gaps = 6/121 (4%)
Query 36 KRKTFEHGGRVWLCTAMGQGTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVL 95
K KT + GR ++ G RK + A + RGTG++ V GR+ + + + +R ++
Sbjct 144 KIKTLDEFGR-----SIAVGKRKSSTAKVFVVRGTGEILVNGRQ-LNDYFLKMKDRESIM 197
Query 96 QPLLLAAAAGLFDVDIQVRGGGPSGQAGAARLAVARALVAACEDCQPALQEAMMLYEDTR 155
PL + + G +++ GGGP+GQA + A+A+ALV + L +A +L D R
Sbjct 198 YPLQVIESVGKYNIFATTSGGGPTGQAESIMHAIAKALVVFNPLLKSRLHKAGVLTRDYR 257
Query 156 Q 156
Sbjct 258 H 258
> SPAC29A4.03c
Length=132
Score = 58.5 bits (140), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 65/127 (51%), Gaps = 3/127 (2%)
Query 51 AMGQGTRKRAAAVALLTRGTGQVRVCGRE-EVYTRWPYVYNRMEVLQPLLLAAAAGLFDV 109
++ +G RK + A + GTG+ V G +VY + + +R + PL ++V
Sbjct 8 SITKGKRKSSKATVKMLPGTGKFYVNGSPFDVY--FQRMVHRKHAVYPLAACNRLTNYNV 65
Query 110 DIQVRGGGPSGQAGAARLAVARALVAACEDCQPALQEAMMLYEDTRQRMPKMPGRMKARK 169
V GGGP+GQ+GA A++++L+ + +++ + D R+ K G+ KARK
Sbjct 66 WATVHGGGPTGQSGAVHAAISKSLILQEPSLKQVIKDTHCVLNDKRKVERKKTGQPKARK 125
Query 170 QRQWSKR 176
+ W KR
Sbjct 126 KYTWVKR 132
> YMR143w
Length=143
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 65/135 (48%), Gaps = 15/135 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQV- 113
G +K A AVA + G G ++V G + P + R +V +PLLL ++DI+V
Sbjct 11 GKKKSATAVAHVKAGKGLIKVNG-SPITLVEPEIL-RFKVYEPLLLVGLDKFSNIDIRVR 68
Query 114 -RGGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYE------DTRQRMPKM 161
GGG Q A R A+A+ LVA E + L++A Y+ D+R+ PK
Sbjct 69 VTGGGHVSQVYAIRQAIAKGLVAYHQKYVDEQSKNELKKAFTSYDRTLLIADSRRPEPKK 128
Query 162 PGRMKARKQRQWSKR 176
G AR + Q S R
Sbjct 129 FGGKGARSRFQKSYR 143
> YDL083c
Length=143
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 65/135 (48%), Gaps = 15/135 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQV- 113
G +K A AVA + G G ++V G + P + R +V +PLLL ++DI+V
Sbjct 11 GKKKSATAVAHVKAGKGLIKVNG-SPITLVEPEIL-RFKVYEPLLLVGLDKFSNIDIRVR 68
Query 114 -RGGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYE------DTRQRMPKM 161
GGG Q A R A+A+ LVA E + L++A Y+ D+R+ PK
Sbjct 69 VTGGGHVSQVYAIRQAIAKGLVAYHQKYVDEQSKNELKKAFTSYDRTLLIADSRRPEPKK 128
Query 162 PGRMKARKQRQWSKR 176
G AR + Q S R
Sbjct 129 FGGKGARSRFQKSYR 143
> SPBC18H10.14
Length=140
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 46/135 (34%), Positives = 69/135 (51%), Gaps = 15/135 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQVR 114
G + A AVA G G ++V G + P + RM+V +P+L+A A VDI+VR
Sbjct 8 GKKGNATAVAHCKVGKGLIKVNG-APLSLVQPEIL-RMKVYEPILVAGADKFAGVDIRVR 65
Query 115 --GGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYE------DTRQRMPKM 161
GGG Q A R A+++A+VA E + L++A++ Y+ D R+ PK
Sbjct 66 VSGGGHVSQIYAIRQAISKAIVAYYQKFVDEHSKAELKKALITYDRTLLVADPRRMEPKK 125
Query 162 PGRMKARKQRQWSKR 176
G AR ++Q S R
Sbjct 126 FGGHGARARQQKSYR 140
> SPAC664.04c
Length=140
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 46/135 (34%), Positives = 69/135 (51%), Gaps = 15/135 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQVR 114
G + A AVA G G ++V G + P + RM+V +P+L+A A VDI+VR
Sbjct 8 GKKGNATAVAHCKVGKGLIKVNG-APLSLVQPEIL-RMKVYEPILVAGADKFAGVDIRVR 65
Query 115 --GGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYE------DTRQRMPKM 161
GGG Q A R A+++A+VA E + L++A++ Y+ D R+ PK
Sbjct 66 VSGGGHVSQIYAIRQAISKAIVAYYQKFVDEHSKAELKKALITYDRTLLVADPRRMEPKK 125
Query 162 PGRMKARKQRQWSKR 176
G AR ++Q S R
Sbjct 126 FGGHGARARQQKSYR 140
> CE12918
Length=144
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 47/135 (34%), Positives = 65/135 (48%), Gaps = 15/135 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQ-- 112
G +K A AVA +G G ++V GR + P + R+++ +PLLL DVDI+
Sbjct 12 GRKKTATAVAHCKKGQGLIKVNGRPLEFLE-PQIL-RIKLQEPLLLVGKERFQDVDIRIR 69
Query 113 VRGGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYE------DTRQRMPKM 161
V GGG Q A R A+A+ALVA E + L+ Y+ D R+R K
Sbjct 70 VSGGGHVAQIYAVRQALAKALVAYYHKYVDEQSKRELKNIFAAYDKSLLVADPRRRESKK 129
Query 162 PGRMKARKQRQWSKR 176
G AR + Q S R
Sbjct 130 FGGPGARARYQKSYR 144
> At3g04230
Length=146
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 58/108 (53%), Gaps = 11/108 (10%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGRE-EVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQV 113
G +K A AV RG+G +++ G E+Y P + R ++ +P+LL VD+++
Sbjct 14 GRKKTATAVTYCKRGSGMIKLNGSPIELY--QPEIL-RFKIFEPVLLLGKHRFAGVDMRI 70
Query 114 R--GGGPSGQAGAARLAVARALVAAC-----EDCQPALQEAMMLYEDT 154
R GGG + + A R ++A+ALVA E + +++ +M Y+ T
Sbjct 71 RATGGGNTSRVYAIRQSIAKALVAYYQKYVDEQSKKEIKDILMRYDRT 118
> Hs20562294
Length=496
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 68/136 (50%), Gaps = 17/136 (12%)
Query 26 AHVHSCFDPEKRKTFEHGGRVWLCTAMGQGTRKRAAAVALLTRGTGQVRVCGREEVYTRW 85
AH ++C P+ +K RV+ G +K A AVA RG G ++V GR
Sbjct 348 AHKNTC-KPQIKKWRRQSVRVF-------GHKKTATAVAHCKRGNGLIKVNGRPLEKIEP 399
Query 86 PYVYNRMEVLQPLLLAAAAGLFDVDIQV--RGGGPSGQAGAARLAVARALVAAC-----E 138
P + + ++L+P+LL + VDI+V GGG Q A R ++++ALVA E
Sbjct 400 PTL--QYKLLEPVLLLSKERFAGVDIRVRVEGGGHVAQIYATRQSISKALVAYYQKYMDE 457
Query 139 DCQPALQEAMMLYEDT 154
+ ++ ++LY+ T
Sbjct 458 ASKKEIKGILILYDWT 473
> At2g09990
Length=146
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 67/135 (49%), Gaps = 15/135 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQVR 114
G +K A AV RG+G +++ G + P + R ++ +P+LL V++++R
Sbjct 14 GRKKTAVAVTHCKRGSGLIKLNG-CPIELFQPEIL-RFKIFEPILLLGKHRFAGVNMRIR 71
Query 115 --GGGPSGQAGAARLAVARALVAAC-----EDCQPALQEAMMLYE------DTRQRMPKM 161
GGG + Q A R ++A+ALVA E + +++ ++ Y+ D R+ PK
Sbjct 72 VNGGGHTSQVYAIRQSIAKALVAYYQKYVDEQSKKEIKDILVRYDRTLLVADPRRCEPKK 131
Query 162 PGRMKARKQRQWSKR 176
G AR + Q S R
Sbjct 132 FGGRGARSRYQKSYR 146
> Hs4506691
Length=146
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 57/109 (52%), Gaps = 13/109 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGR--EEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQ 112
G +K A AVA RG G ++V GR E + R + ++L+P+LL VDI+
Sbjct 14 GRKKTATAVAHCKRGNGLIKVNGRPLEMIEPRT----LQYKLLEPVLLLGKERFAGVDIR 69
Query 113 V--RGGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYEDT 154
V +GGG Q A R ++++ALVA E + +++ ++ Y+ T
Sbjct 70 VRVKGGGHVAQIYAIRQSISKALVAYYQKYVDEASKKEIKDILIQYDRT 118
> At5g18380
Length=146
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 67/135 (49%), Gaps = 15/135 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQVR 114
G +K A AV RG+G +++ G + P + R ++ +P+LL V++++R
Sbjct 14 GRKKTAVAVTHCKRGSGLIKLNG-CPIELFQPEIL-RFKIFEPVLLLGKHRFAGVNMRIR 71
Query 115 --GGGPSGQAGAARLAVARALVAAC-----EDCQPALQEAMMLYE------DTRQRMPKM 161
GGG + Q A R ++A+ALVA E + +++ ++ Y+ D R+ PK
Sbjct 72 VNGGGHTSQVYAIRQSIAKALVAYYQKYVDEQSKKEIKDILVRYDRTLLVADPRRCEPKK 131
Query 162 PGRMKARKQRQWSKR 176
G AR + Q S R
Sbjct 132 FGGRGARSRYQKSYR 146
> 7291436
Length=148
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 46/137 (33%), Positives = 67/137 (48%), Gaps = 19/137 (13%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGR--EEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQ 112
G +K A AVA RG G ++V GR E++ P V ++ +PLLL VDI+
Sbjct 16 GRKKTATAVAYCKRGNGLLKVNGRPLEQI---EPKVLQ-YKLQEPLLLLGKEKFAGVDIR 71
Query 113 V--RGGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYE------DTRQRMP 159
V GGG Q A R A+++ALVA E + +++ ++ Y+ D R+ P
Sbjct 72 VRVSGGGHVAQIYAIRQAISKALVAFYQKYVDEASKKEIKDILVQYDRTLLVGDPRRCEP 131
Query 160 KMPGRMKARKQRQWSKR 176
K G AR + Q S R
Sbjct 132 KKFGGPGARARYQKSYR 148
> Hs18549572
Length=146
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 56/112 (50%), Gaps = 19/112 (16%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPY--VYNRM---EVLQPLLLAAAAGLFDV 109
G +K A AVA RG G ++V G WP + RM ++L+P+LL V
Sbjct 14 GRKKTATAVAHCKRGNGLIKVNG-------WPLEMIELRMLQYKLLEPVLLLGKERFAGV 66
Query 110 DI--QVRGGGPSGQAGAARLAVARALVAAC-----EDCQPALQEAMMLYEDT 154
DI +++GGG Q A ++++ALVA E + +++ ++ Y+ T
Sbjct 67 DICVRMKGGGHVAQIYAIHQSISKALVAYYQKYVDEASKKEIKDILIQYDQT 118
> Hs18591367
Length=146
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 54/107 (50%), Gaps = 9/107 (8%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQV- 113
G +K A AVA RG G ++V G+ T + + E+L+P+LL VDI+V
Sbjct 14 GRKKTATAVAHCRRGNGLIKVNGQPLEMTEPRML--QYELLEPVLLLGKERFAGVDIRVR 71
Query 114 -RGGGPSGQAGAARLAVARALVA-----ACEDCQPALQEAMMLYEDT 154
+GG Q A R + ++ALVA E + +++ ++ Y+ T
Sbjct 72 VKGGAHVAQIYAIRQSTSKALVAYYQKYVDEASKKEIKDILIQYDRT 118
> Hs7305013
Length=351
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 27/56 (48%), Gaps = 3/56 (5%)
Query 3 EETQTNSAEPAAAAAAAADGLLRAHVHSCFDPEKRKTFEHGGRVWLCTAMGQGTRK 58
E + E A A+ L+ A V+SC D E R+TF R+ C + Q TR+
Sbjct 269 ESCNVLAVEKYFLLLAEANSLVNAAVYSCRDAEMRRTFR---RLLCCACLRQSTRE 321
> ECU03g0310
Length=145
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 38/142 (26%), Positives = 66/142 (46%), Gaps = 26/142 (18%)
Query 54 QGTRKRAAAVALLTR-GTGQVRVCGREEVYTRWPY--VYNRMEV--LQPLL-LAAAAGLF 107
QG +KRA A + T+ G +R+ + P+ + N + + L+ ++ + A +
Sbjct 11 QGKKKRAVATCVCTQTGEFSIRI-------NKVPFKLITNTLMIAKLKEIICIVEQANIK 63
Query 108 DV--DIQVRGGGPSGQAGAARLAVARALVAAC---------EDCQPALQEA--MMLYEDT 154
D+ DI + GG + A R+A A+A++A ++ Q L + L D+
Sbjct 64 DLSFDITCKIGGDVSRVYAVRMAFAKAILAFYGTYFDEWKKQEIQKRLLDFDRFSLVADS 123
Query 155 RQRMPKMPGRMKARKQRQWSKR 176
R+R PK G AR + Q S R
Sbjct 124 RKREPKKFGGPGARARYQKSYR 145
> Hs18552340
Length=132
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 9/81 (11%)
Query 55 GTRKRAAAVALLTRGTGQVRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQVR 114
G +KRA AVA G ++V G + + ++L+PLLL A + L
Sbjct 14 GCKKRATAVAPGKHSNGLIKVNGWHLEMIELHML--QYKLLEPLLLWAKSDLL------- 64
Query 115 GGGPSGQAGAARLAVARALVA 135
GG Q A ++++ALVA
Sbjct 65 GGSHVAQICAVHQSISKALVA 85
> Hs18152783
Length=214
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 23/101 (22%), Positives = 44/101 (43%), Gaps = 6/101 (5%)
Query 73 VRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQVRGGGPSGQAGAARLAVARA 132
V+ CGR+ + R + V++ + + AG + +RG Q AR+ + +
Sbjct 77 VKSCGRDGFHMR--VRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQV 134
Query 133 LVAACEDCQPALQEAMMLYEDTRQRMPKMPGRMKARKQRQW 173
+++ + LQ + E R+ K PGR K ++W
Sbjct 135 IMSI----RTKLQNEEHVIEALRRAKFKFPGRQKIHISKKW 171
> At2g17790
Length=830
Score = 28.5 bits (62), Expect = 7.7, Method: Composition-based stats.
Identities = 29/92 (31%), Positives = 40/92 (43%), Gaps = 7/92 (7%)
Query 4 ETQTNSAEPAAAAAAAADGLLRAHVHSC---FDPEKRKTFEHGGRVWLCTAMGQGTRKRA 60
+T T+ A AA RA V++C F E R+T + G RV LC A
Sbjct 683 DTLTHKATGYAAKLLKKPDQCRA-VYACSHLFWLEDRETIQDGERVLLCLKRALKIANSA 741
Query 61 AAVALLTRG-TGQVRVCGREEVYTRWPYVYNR 91
VA RG TG V + E+ ++ Y Y +
Sbjct 742 QQVANTARGSTGSVTLF--IEILNKYLYFYEK 771
> Hs5174431
Length=214
Score = 28.1 bits (61), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 22/101 (21%), Positives = 44/101 (43%), Gaps = 6/101 (5%)
Query 73 VRVCGREEVYTRWPYVYNRMEVLQPLLLAAAAGLFDVDIQVRGGGPSGQAGAARLAVARA 132
V+ CG++ + R + V++ + + AG + +RG Q AR+ + +
Sbjct 77 VKSCGKDGFHIR--VRLHPFHVIRINKMLSCAGADRLQTGMRGAFGKPQGTVARVHIGQV 134
Query 133 LVAACEDCQPALQEAMMLYEDTRQRMPKMPGRMKARKQRQW 173
+++ + LQ + E R+ K PGR K ++W
Sbjct 135 IMSI----RTKLQNKEHVIEALRRAKFKFPGRQKIHISKKW 171
Lambda K H
0.320 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40