bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4022_orf2
Length=139
Score E
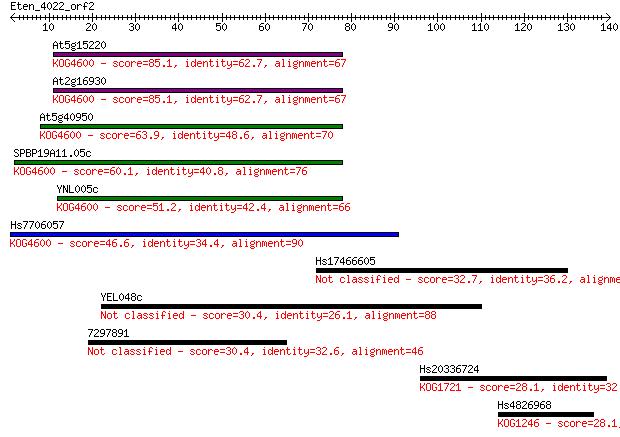
Sequences producing significant alignments: (Bits) Value
At5g15220 85.1 4e-17
At2g16930 85.1 4e-17
At5g40950 63.9 8e-11
SPBP19A11.05c 60.1 1e-09
YNL005c 51.2 6e-07
Hs7706057 46.6 2e-05
Hs17466605 32.7 0.23
YEL048c 30.4 1.0
7297891 30.4 1.1
Hs20336724 28.1 4.8
Hs4826968 28.1 5.1
> At5g15220
Length=162
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 42/67 (62%), Positives = 48/67 (71%), Gaps = 0/67 (0%)
Query 11 WATSKGGSATKNNRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAGEGVMRGGDDNLV 70
WAT K +TKN RDS PKFLGVKKFGG+ V+PG+I+VRQRGTRF G+ V G D L
Sbjct 46 WATKKTAGSTKNGRDSNPKFLGVKKFGGESVIPGNIIVRQRGTRFHPGDYVGIGKDHTLF 105
Query 71 ALNSGFV 77
AL G V
Sbjct 106 ALKEGRV 112
> At2g16930
Length=147
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 42/67 (62%), Positives = 48/67 (71%), Gaps = 0/67 (0%)
Query 11 WATSKGGSATKNNRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAGEGVMRGGDDNLV 70
WAT K +TKN RDS PKFLGVKKFGG+ V+PG+I+VRQRGTRF G+ V G D L
Sbjct 39 WATKKTAGSTKNGRDSNPKFLGVKKFGGESVIPGNIIVRQRGTRFHPGDYVGIGKDHTLF 98
Query 71 ALNSGFV 77
AL G V
Sbjct 99 ALKEGRV 105
> At5g40950
Length=198
Score = 63.9 bits (154), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 34/70 (48%), Positives = 43/70 (61%), Gaps = 0/70 (0%)
Query 8 TVCWATSKGGSATKNNRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAGEGVMRGGDD 67
T+ A KG +TKN RDS + LGVK +G Q PG I+VRQRGT+F AG+ V G D
Sbjct 55 TIESAHKKGAGSTKNGRDSPGQRLGVKIYGDQVAKPGAIIVRQRGTKFHAGKNVGIGKDH 114
Query 68 NLVALNSGFV 77
+ +L G V
Sbjct 115 TIFSLIDGLV 124
> SPBP19A11.05c
Length=153
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/76 (40%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 2 NSCTSATVCWATSKGGSATKNNRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAGEGV 61
N T + +T GG ++KN DS + LG+K+ QFV G+IL+RQRGT+F G+
Sbjct 27 NILTFPPIRTSTKHGGGSSKNTGDSAGRRLGIKRSENQFVRAGEILIRQRGTKFHPGDNT 86
Query 62 MRGGDDNLVALNSGFV 77
G D + +L SG+V
Sbjct 87 GLGKDHTIYSLVSGYV 102
> YNL005c
Length=371
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/66 (42%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 12 ATSKGGSATKNNRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAGEGVMRGGDDNLVA 71
AT + + + +DS + LG KK+ GQ V G+I++RQRGT+F GE V G D ++ A
Sbjct 28 ATKRAAGSRTSMKDSAGRRLGPKKYEGQDVSTGEIIMRQRGTKFYPGENVGIGKDHSIFA 87
Query 72 LNSGFV 77
L G V
Sbjct 88 LEPGVV 93
> Hs7706057
Length=148
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 45/92 (48%), Gaps = 2/92 (2%)
Query 1 PNSCTSATVCWATSKGGSATKN-NRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAGE 59
P T+ V +A+ K G ++KN S + G+KK G +V G+I+ QR R+ G
Sbjct 20 PTPATALAVRYASKKSGGSSKNLGGKSSGRRQGIKKMEGHYVHAGNIIATQRHFRWHPGA 79
Query 60 GVMRGGDDNLVALNSGFV-FMRRVYRTSDANT 90
V G + L AL G V + + VY NT
Sbjct 80 HVGVGKNKCLYALEEGIVRYTKEVYVPHPRNT 111
> Hs17466605
Length=170
Score = 32.7 bits (73), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query 72 LNSGFVFMRRVYRTSDANTRIKAGCVVSIKASPAETLDPKAARVSFLRTLAKSWMMQG 129
+N FVF + Y +T++ C+V+ K S ET K A V++ TLA S QG
Sbjct 1 MNISFVFFYQKYHQLILSTKVAILCIVTRKLSKNET---KTAGVTYEATLANSGFPQG 55
> YEL048c
Length=152
Score = 30.4 bits (67), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 23/90 (25%), Positives = 40/90 (44%), Gaps = 4/90 (4%)
Query 22 NNRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAG--EGVMRGGDDNLVALNSGFVFM 79
++ DS+P + + G V +L++Q G + G + + G DD A+N F +
Sbjct 55 HSLDSKPLLKSIFQLEGVSVFA--MLIKQTGLKIVIGFEQKSLSGADDEFEAINQIFETV 112
Query 80 RRVYRTSDANTRIKAGCVVSIKASPAETLD 109
R++Y N + +G SI S D
Sbjct 113 RKIYIRVKCNPLLVSGDEKSIIKSLERKFD 142
> 7297891
Length=690
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 19 ATKNNRDSRPKFLGVKKFGGQFVLPGDILVRQRGTRFKAGEGVMRG 64
A +N+R + PKFL + +F G F D +V G+ +RG
Sbjct 110 AGRNSRINSPKFLRLTQFAGNFSDAFDFVVVNNGSSASGNYRTLRG 155
> Hs20336724
Length=665
Score = 28.1 bits (61), Expect = 4.8, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 19/43 (44%), Gaps = 0/43 (0%)
Query 96 CVVSIKASPAETLDPKAARVSFLRTLAKSWMMQGQPLELPQQQ 138
C S + + +PKA LR +W Q +P ELP Q
Sbjct 47 CKPSCVSQLGQRAEPKATERGILRATGVAWESQLKPEELPSMQ 89
> Hs4826968
Length=1722
Score = 28.1 bits (61), Expect = 5.1, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 114 RVSFLRTLAKSWMMQGQPLELP 135
R+ FL LAK W +QG L++P
Sbjct 87 RLDFLDQLAKFWELQGSTLKIP 108
Lambda K H
0.319 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40