bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
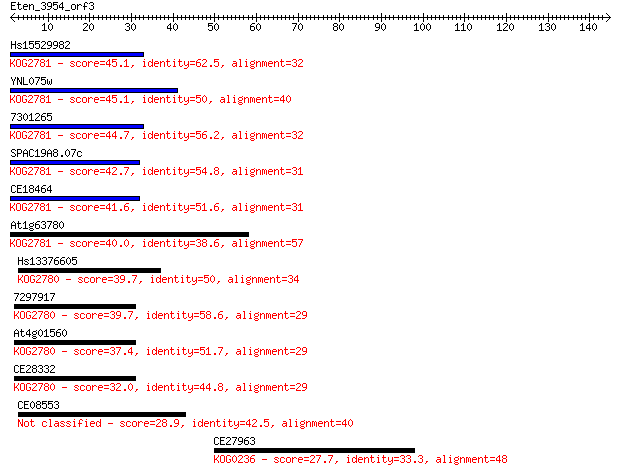
Query= Eten_3954_orf3
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
Hs15529982 45.1 5e-05
YNL075w 45.1 5e-05
7301265 44.7 5e-05
SPAC19A8.07c 42.7 2e-04
CE18464 41.6 5e-04
At1g63780 40.0 0.002
Hs13376605 39.7 0.002
7297917 39.7 0.002
At4g01560 37.4 0.009
CE28332 32.0 0.44
CE08553 28.9 3.5
CE27963 27.7 7.0
> Hs15529982
Length=291
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPE 32
G P LIV HLP GPTA+FTL V +RHD+P+
Sbjct 144 GTPVGLIVSHLPFGPTAYFTLCNVVMRHDIPD 175
> YNL075w
Length=290
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 20/40 (50%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPNMSQV 40
G P L + H PHGPTA F+L V +RHD+ A N S+V
Sbjct 147 GVPTSLTISHFPHGPTAQFSLHNVVMRHDIIN-AGNQSEV 185
> 7301265
Length=298
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPE 32
G P L+V HLP+GPTA F +S V +RHD+P+
Sbjct 142 GIPDSLVVCHLPYGPTAFFNISDVVMRHDIPD 173
> SPAC19A8.07c
Length=289
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 17/31 (54%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLP 31
G P L++ HLP+GPT F+L V LRHD+P
Sbjct 145 GIPDGLVISHLPYGPTLSFSLHNVVLRHDIP 175
> CE18464
Length=292
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLP 31
G P ++V HLP GPTA F+++ V +RHD+P
Sbjct 139 GNPDGMLVCHLPFGPTAFFSMANVVMRHDIP 169
> At1g63780
Length=294
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 31/66 (46%), Gaps = 12/66 (18%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHD---------LPERAPNMSQVLFCCCCSAAAA 51
G P LI+ HLP GPTA+F L V RHD +PE+ P++ +F +
Sbjct 144 GVPDGLIISHLPFGPTAYFGLLNVVTRHDISDKKSIGKMPEQYPHL---IFNNFTTQMGQ 200
Query 52 AAAALL 57
+L
Sbjct 201 RVGNIL 206
> Hs13376605
Length=349
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 3 PPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPN 36
P LI+ HLP+GPTAHF +S V LR ++ R +
Sbjct 205 PNGLILSHLPNGPTAHFKMSSVRLRKEIKRRGKD 238
> 7297917
Length=394
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/29 (58%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 2 QPPRLIVFHLPHGPTAHFTLSGVCLRHDL 30
+P L+V HLP+GPTAHF LS V L D+
Sbjct 247 KPNGLLVIHLPNGPTAHFKLSNVKLTSDI 275
> At4g01560
Length=343
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 2 QPPRLIVFHLPHGPTAHFTLSGVCLRHDL 30
+P L++ LP+GPTAHF LS + LR D+
Sbjct 189 EPDALLIIGLPNGPTAHFKLSNLVLRKDI 217
> CE28332
Length=384
Score = 32.0 bits (71), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 1/30 (3%)
Query 2 QPPRLIVF-HLPHGPTAHFTLSGVCLRHDL 30
+ P I+F HLP GPTA+F ++ + DL
Sbjct 238 KKPNGIIFCHLPEGPTAYFKINSLTFTQDL 267
> CE08553
Length=461
Score = 28.9 bits (63), Expect = 3.5, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 22/40 (55%), Gaps = 3/40 (7%)
Query 3 PPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPNMSQVLF 42
P L +F LP P+AH L+G + H P +PNM V F
Sbjct 115 PLCLKLFSLPFAPSAH--LTGPFISH-YPHYSPNMDAVSF 151
> CE27963
Length=652
Score = 27.7 bits (60), Expect = 7.0, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 50 AAAAAALLLLPFWRLLRAAPVCLFAAAAAVLLLLECRCCRELPPLLRC 97
+A AL++ L + P C+ +A V+L R C LP L RC
Sbjct 393 SACFMALVITTIGPYLASLPSCILSAIVIVVLESMLRKCTVLPGLWRC 440
Lambda K H
0.330 0.141 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1675978996
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40