bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
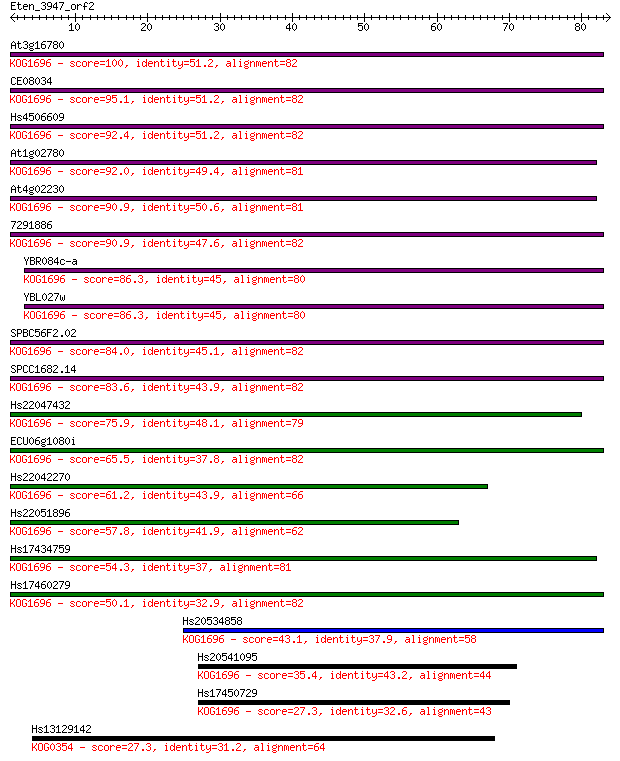
Query= Eten_3947_orf2
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
At3g16780 100 6e-22
CE08034 95.1 3e-20
Hs4506609 92.4 2e-19
At1g02780 92.0 2e-19
At4g02230 90.9 5e-19
7291886 90.9 6e-19
YBR084c-a 86.3 1e-17
YBL027w 86.3 1e-17
SPBC56F2.02 84.0 6e-17
SPCC1682.14 83.6 7e-17
Hs22047432 75.9 2e-14
ECU06g1080i 65.5 3e-11
Hs22042270 61.2 4e-10
Hs22051896 57.8 5e-09
Hs17434759 54.3 5e-08
Hs17460279 50.1 9e-07
Hs20534858 43.1 1e-04
Hs20541095 35.4 0.026
Hs17450729 27.3 6.5
Hs13129142 27.3 7.5
> At3g16780
Length=209
Score = 100 bits (249), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 42/82 (51%), Positives = 60/82 (73%), Gaps = 0/82 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
+KCG+G+ WLDPNE+GDI+MANSR ++R+L++D IIR+ + S+ R R +AKR+GR
Sbjct 15 MKCGKGKVWLDPNESGDISMANSRQNIRKLVKDGFIIRKPTKIHSRSRARALNEAKRKGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
H R+ T +A LP K+LWM
Sbjct 75 HSGYGKRKGTREARLPTKILWM 96
> CE08034
Length=198
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 42/82 (51%), Positives = 58/82 (70%), Gaps = 0/82 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
LKCG+ R WLDPNE +I+ ANSR S+R L+ D LIIR+ V V S++R R Y++A+R+GR
Sbjct 15 LKCGKHRVWLDPNEVSEISGANSRQSIRRLVNDGLIIRKPVTVHSRFRAREYEEARRKGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
H R+ T +A +P K LW+
Sbjct 75 HTGYGKRRGTANARMPEKTLWI 96
> Hs4506609
Length=196
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 42/82 (51%), Positives = 57/82 (69%), Gaps = 0/82 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L+CG+ + WLDPNE +IA ANSR +R+L++D LIIR+ V V S+ R R A+R+GR
Sbjct 15 LRCGKKKVWLDPNETNEIANANSRQQIRKLIKDGLIIRKPVTVHSRARCRKNTLARRKGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
HM I R+ T +A +P KV WM
Sbjct 75 HMGIGKRKGTANARMPEKVTWM 96
> At1g02780
Length=214
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 57/81 (70%), Gaps = 0/81 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
+KCG+G+ WLDPNE+ DI+MANSR ++R+L++D IIR+ + S+ R R + AK +GR
Sbjct 15 MKCGKGKVWLDPNESSDISMANSRQNIRKLVKDGFIIRKPTKIHSRSRARKMKIAKMKGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLW 81
H R+ T +A LP KVLW
Sbjct 75 HSGYGKRKGTREARLPTKVLW 95
> At4g02230
Length=208
Score = 90.9 bits (224), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 55/81 (67%), Gaps = 0/81 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
LKCG+ + WLDPNE DI+MANSR ++R+L++D IIR+ + S+ R R AKR+GR
Sbjct 15 LKCGKRKVWLDPNEGSDISMANSRQNIRKLVKDGFIIRKPTKIHSRSRARQLNIAKRKGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLW 81
H R+ T +A LP KVLW
Sbjct 75 HSGYGKRKGTREARLPTKVLW 95
> 7291886
Length=203
Score = 90.9 bits (224), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 39/82 (47%), Positives = 58/82 (70%), Gaps = 0/82 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L+CG+ + WLDPNE +IA NSR ++R+L++D LII++ V V S+YR R +A+R+GR
Sbjct 15 LRCGKKKVWLDPNEINEIANTNSRQNIRKLIKDGLIIKKPVVVHSRYRVRKNTEARRKGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
H R+ T +A +P K+LWM
Sbjct 75 HCGFGKRKGTANARMPTKLLWM 96
> YBR084c-a
Length=189
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 57/80 (71%), Gaps = 0/80 (0%)
Query 3 CGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGRHM 62
G+ + WLDPNE +IA ANSR ++R+L+++ I+++AV V SK R R + Q+KR+GRH
Sbjct 17 VGKRKVWLDPNETSEIAQANSRNAIRKLVKNGTIVKKAVTVHSKSRTRAHAQSKREGRHS 76
Query 63 CIAMRQVTLDAPLPAKVLWM 82
R+ T +A LP++V+W+
Sbjct 77 GYGKRKGTREARLPSQVVWI 96
> YBL027w
Length=189
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 57/80 (71%), Gaps = 0/80 (0%)
Query 3 CGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGRHM 62
G+ + WLDPNE +IA ANSR ++R+L+++ I+++AV V SK R R + Q+KR+GRH
Sbjct 17 VGKRKVWLDPNETSEIAQANSRNAIRKLVKNGTIVKKAVTVHSKSRTRAHAQSKREGRHS 76
Query 63 CIAMRQVTLDAPLPAKVLWM 82
R+ T +A LP++V+W+
Sbjct 77 GYGKRKGTREARLPSQVVWI 96
> SPBC56F2.02
Length=193
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 37/82 (45%), Positives = 56/82 (68%), Gaps = 0/82 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
LKCG+ + W+DPNE +I+ ANSR +VR+L++D L+IR+ + S++R R AKR GR
Sbjct 15 LKCGKRKVWMDPNEISEISNANSRQNVRKLIKDGLVIRKPNLMHSRFRIRKTHAAKRLGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
H R+ T +A +P+ V+WM
Sbjct 75 HTGYGKRKGTAEARMPSAVVWM 96
> SPCC1682.14
Length=193
Score = 83.6 bits (205), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 36/82 (43%), Positives = 56/82 (68%), Gaps = 0/82 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
LKCG+ + W+DPNE +I+ ANSR ++R+L++D L+IR+ + S++R R AKR GR
Sbjct 15 LKCGKRKVWMDPNEISEISNANSRQNIRKLVKDGLVIRKPNLMHSRFRIRKTHAAKRLGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
H R+ T +A +P+ V+WM
Sbjct 75 HTGYGKRKGTAEARMPSTVVWM 96
> Hs22047432
Length=521
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/79 (48%), Positives = 53/79 (67%), Gaps = 0/79 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L+ G+ + WLDPNEA +IA ANSR +R+L++D LII + V V S+ + R A R+GR
Sbjct 15 LRYGKKKVWLDPNEANEIASANSRQQIRKLIKDGLIICKPVTVHSQAQCRKNTLAHRKGR 74
Query 61 HMCIAMRQVTLDAPLPAKV 79
HM R+ T +AP+P KV
Sbjct 75 HMGTGKRKGTANAPMPEKV 93
> ECU06g1080i
Length=171
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 50/82 (60%), Gaps = 0/82 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L CG+ + WLD NE + A++R +++L++DN+II + S+ R R +AK++GR
Sbjct 15 LNCGKKKLWLDNNEIERLNGASTREQIKQLIKDNVIIMKQDKHTSRGRCRKRLEAKKKGR 74
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
HM R T +A +P K +WM
Sbjct 75 HMGTGKRFGTANARMPQKKIWM 96
> Hs22042270
Length=383
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L CG+ + WLDPNE +I+ ANS + +L++D L+IR+ + V S+ R + A R+GR
Sbjct 259 LPCGKKKVWLDPNETNEISNANSCQQIWKLIKDGLMIRKPMTVHSQARCQENTLAYRKGR 318
Query 61 HMCIAM 66
HM IA+
Sbjct 319 HMGIAI 324
> Hs22051896
Length=145
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L+CG+ + WLDP E +IA ANS +R+L++D LII + +D R Y ++K+ R
Sbjct 15 LRCGKKKVWLDPKETSEIANANSHQWIRKLIKDGLIIHKP-KIDHHMYHRRYHESKKVDR 73
Query 61 HM 62
HM
Sbjct 74 HM 75
> Hs17434759
Length=138
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 45/81 (55%), Gaps = 8/81 (9%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L CG+ + WLDP E +IA NSR +R+L+RD LII + V V S+ + A R+
Sbjct 15 LCCGKKKVWLDPGETSEIANVNSRQQIRKLIRDGLIILKPVPVHSQAQCWKNTLAGRK-- 72
Query 61 HMCIAMRQVTLDAPLPAKVLW 81
++ T +A +P K+ W
Sbjct 73 ------QKGTANARMPEKITW 87
> Hs17460279
Length=170
Score = 50.1 bits (118), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 39/82 (47%), Gaps = 26/82 (31%)
Query 1 LKCGRGRFWLDPNEAGDIAMANSRFSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGR 60
L+CG+ + WLDPN+ +IA ANS + +L++D LIIR+ V
Sbjct 15 LRCGKKKVWLDPNKTNEIANANSHQQIWKLIKDGLIIRKPVT------------------ 56
Query 61 HMCIAMRQVTLDAPLPAKVLWM 82
T +A +P KV WM
Sbjct 57 --------GTANARMPEKVTWM 70
> Hs20534858
Length=376
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/58 (37%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 25 FSVRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGRHMCIAMRQVTLDAPLPAKVLWM 82
+++L++D LII + V V S+ + A R+GRHM I ++ T +A +P KV WM
Sbjct 251 LEIQKLIKDGLIIHKPVIVHSQAQCWKNTLACRKGRHMGIGKQKRTANAQIPEKVTWM 308
> Hs20541095
Length=240
Score = 35.4 bits (80), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Query 27 VRELLRDNLIIREAVAVDSKYRFRLYQQAKRQGRHMCIAMRQVT 70
+R+L++D LII + V V S+ +F A ++G+HMCI QVT
Sbjct 97 IRKLIKDGLIIWKPVTVHSQAQFWKNTLAHQKGKHMCIG--QVT 138
> Hs17450729
Length=147
Score = 27.3 bits (59), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 28/45 (62%), Gaps = 2/45 (4%)
Query 27 VRELLRDNLIIREAVAVDSKYRFRLYQQ--AKRQGRHMCIAMRQV 69
+R+L++D LI+R V R + ++ A+R+GRH+ I +++
Sbjct 22 IRKLIKDGLILRHRKPVTVHSRAQCWKSTLARRKGRHLGIESKKI 66
> Hs13129142
Length=678
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 29/66 (43%), Gaps = 2/66 (3%)
Query 4 GRGRFWLDPNEAGDIAMANSRFSVRELLRDNL--IIREAVAVDSKYRFRLYQQAKRQGRH 61
RGR W D + +A SR REL+ + L ++ +AVA K YQ R +
Sbjct 468 ARGRAWADQSVYAFVATEGSRELKRELINEALETLMEQAVAAVQKMDQAEYQAKIRDLQQ 527
Query 62 MCIAMR 67
+ R
Sbjct 528 AALTKR 533
Lambda K H
0.329 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200681374
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40