bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3942_orf3
Length=210
Score E
Sequences producing significant alignments: (Bits) Value
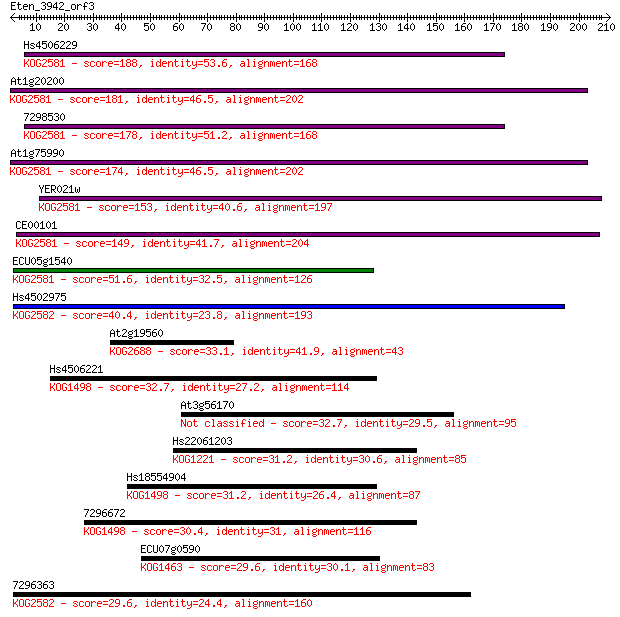
Hs4506229 188 7e-48
At1g20200 181 1e-45
7298530 178 7e-45
At1g75990 174 1e-43
YER021w 153 2e-37
CE00101 149 4e-36
ECU05g1540 51.6 1e-06
Hs4502975 40.4 0.002
At2g19560 33.1 0.38
Hs4506221 32.7 0.53
At3g56170 32.7 0.55
Hs22061203 31.2 1.7
Hs18554904 31.2 1.7
7296672 30.4 3.0
ECU07g0590 29.6 4.3
7296363 29.6 4.6
> Hs4506229
Length=534
Score = 188 bits (477), Expect = 7e-48, Method: Compositional matrix adjust.
Identities = 90/168 (53%), Positives = 127/168 (75%), Gaps = 0/168 (0%)
Query 6 KKCIVVELLMGDIPERPLFNKQNMHAELVPYREIVLAVRSGDLHAFASVLQQHEQRFRLD 65
K IVVELL+G+IP+R F + ++ L+PY + AVR+G+L F VL Q ++F+ D
Sbjct 330 KLLIVVELLLGEIPDRLQFRQPSLKRSLMPYFLLTQAVRTGNLAKFNQVLDQFGEKFQAD 389
Query 66 DTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGKAILDRVIDA 125
TYTL+ RL NVI+ G+R+ISLSYSRI L DIA+KL LDS ++AE + KAI D VI+A
Sbjct 390 GTYTLIIRLRHNVIKTGVRMISLSYSRISLADIAQKLQLDSPEDAEFIVAKAIRDGVIEA 449
Query 126 SIDHDKQFLQSKASVDLYASNEPQKAFHRRITFCLDLYNDAVKAMQYP 173
SI+H+K ++QSK +D+Y++ EPQ AFH+RI+FCLD++N +VKAM++P
Sbjct 450 SINHEKGYVQSKEMIDIYSTREPQLAFHQRISFCLDIHNMSVKAMRFP 497
> At1g20200
Length=488
Score = 181 bits (458), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 94/208 (45%), Positives = 139/208 (66%), Gaps = 6/208 (2%)
Query 1 RLAALKKCIVVELLMGDIPERPLFNKQNMHAELVPYREIVLAVRSGDLHAFASVLQQHEQ 60
R+ K I+V LL+G+IPER +F ++ M L PY E+ AVR GDL F +V ++
Sbjct 278 RIQCNKWAILVRLLLGEIPERSIFTQKGMEKALRPYFELTNAVRIGDLELFRTVQEKFLD 337
Query 61 RFRLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDS---AKEAEAVAGKA 117
F D T+ L+ RL NVIR GLR IS+SYSRI L D+AKKL L+S +AE++ KA
Sbjct 338 TFAQDRTHNLIVRLRHNVIRTGLRNISISYSRISLPDVAKKLRLNSENPVADAESIVAKA 397
Query 118 ILDRVIDASIDHDKQFLQSKASVDLYASNEPQKAFHRRITFCLDLYNDAVKAMQYPGSTE 177
I D IDA+IDH + SK + D+Y++NEPQ AF+ RI FCL+++N+AV+A+++P +T
Sbjct 398 IRDGAIDATIDHKNGCMVSKETGDIYSTNEPQTAFNSRIAFCLNMHNEAVRALRFPPNTH 457
Query 178 RQQTAEDDERKRAMQEE---RARAEDED 202
+++ +++ R+R QEE + AE++D
Sbjct 458 KEKESDEKRRERKQQEEELAKHMAEEDD 485
> 7298530
Length=494
Score = 178 bits (451), Expect = 7e-45, Method: Compositional matrix adjust.
Identities = 86/168 (51%), Positives = 121/168 (72%), Gaps = 0/168 (0%)
Query 6 KKCIVVELLMGDIPERPLFNKQNMHAELVPYREIVLAVRSGDLHAFASVLQQHEQRFRLD 65
K IVVELL+G+IPER +F + + L Y ++ AVR G+L F V+ Q+ +F+LD
Sbjct 291 KLIIVVELLLGNIPERVVFRQAGLRQSLGAYFQLTQAVRLGNLKRFGDVVSQYGPKFQLD 350
Query 66 DTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGKAILDRVIDA 125
T+TL+ RL NVI+ +R I LSYSRI +DIAK+L LDSA++AE + KAI D VI+A
Sbjct 351 HTFTLIIRLRHNVIKTAIRSIGLSYSRISPQDIAKRLMLDSAEDAEFIVSKAIRDGVIEA 410
Query 126 SIDHDKQFLQSKASVDLYASNEPQKAFHRRITFCLDLYNDAVKAMQYP 173
++D + F++SK S D+Y++ EPQ AFH RI+FCL+L+N +VKAM+YP
Sbjct 411 TLDPAQNFMRSKESTDIYSTREPQLAFHERISFCLNLHNQSVKAMRYP 458
> At1g75990
Length=487
Score = 174 bits (441), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 94/208 (45%), Positives = 140/208 (67%), Gaps = 6/208 (2%)
Query 1 RLAALKKCIVVELLMGDIPERPLFNKQNMHAELVPYREIVLAVRSGDLHAFASVLQQHEQ 60
R+ K I+V LL+G+IPER +F ++ M L PY E+ AVR GDL F + ++ +
Sbjct 277 RIQCNKWAIIVRLLLGEIPERSIFTQKGMEKTLRPYFELTNAVRIGDLELFGKIQEKFAK 336
Query 61 RFRLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAK---EAEAVAGKA 117
F D T+ L+ RL NVIR GLR IS+SYSRI L+D+A+KL L+SA +AE++ KA
Sbjct 337 TFAEDRTHNLIVRLRHNVIRTGLRNISISYSRISLQDVAQKLRLNSANPVADAESIVAKA 396
Query 118 ILDRVIDASIDHDKQFLQSKASVDLYASNEPQKAFHRRITFCLDLYNDAVKAMQYPGSTE 177
I D IDA+IDH + SK + D+Y++NEPQ AF+ RI FCL+++N+AV+A+++P +T
Sbjct 397 IRDGAIDATIDHKNGCMVSKETGDIYSTNEPQTAFNSRIAFCLNMHNEAVRALRFPPNTH 456
Query 178 RQQTAEDDERKRAMQEE---RARAEDED 202
R++ +E+ R+ QEE + AE++D
Sbjct 457 REKESEEKRREMKQQEEELAKYMAEEDD 484
> YER021w
Length=523
Score = 153 bits (386), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 80/197 (40%), Positives = 117/197 (59%), Gaps = 5/197 (2%)
Query 11 VELLMGDIPERPLFNKQNMHAELVPYREIVLAVRSGDLHAFASVLQQHEQRFRLDDTYTL 70
++LLMGDIPE F++ NM L+PY + AV+ GDL F S + +++Q DDTY L
Sbjct 320 IQLLMGDIPELSFFHQSNMQKSLLPYYHLTKAVKLGDLKKFTSTITKYKQLLLKDDTYQL 379
Query 71 LCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGKAILDRVIDASIDHD 130
RL NVI+ G+RIISL+Y +I L+DI KL LDS + E + +AI D VI+A I+H+
Sbjct 380 CVRLRSNVIKTGIRIISLTYKKISLRDICLKLNLDSEQTVEYMVSRAIRDGVIEAKINHE 439
Query 131 KQFLQSKASVDLYASNEPQKAFHRRITFCLDLYNDAVKAMQYPGSTERQQTAEDDERKRA 190
F+++ +++Y S +PQ+ F RI F L+++ + +M+YP + QQ K
Sbjct 440 DGFIETTELLNIYDSEDPQQVFDERIKFANQLHDEYLVSMRYPEDKKTQQN-----EKSE 494
Query 191 MQEERARAEDEDLGDDM 207
E D DL DDM
Sbjct 495 NGENDDDTLDGDLMDDM 511
> CE00101
Length=504
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 85/208 (40%), Positives = 121/208 (58%), Gaps = 4/208 (1%)
Query 3 AALKKCIVVELLMGDIPERPLFNKQNMHAELVPYREIVLAVRSGDLHAFASVLQQHEQRF 62
A K +V+ LL G+IP+R +F + L Y ++ VR GD+ F L+Q + +F
Sbjct 295 AVQKWVVVIGLLQGEIPDRSVFRQPIYRKCLAHYLDLSRGVRDGDVARFNHNLEQFKTQF 354
Query 63 RLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGKAILDRV 122
DDT TL+ RL QNVI+ ++ ISL+YSRIY+KDIAKKL + + E E + KAI D
Sbjct 355 EADDTLTLIVRLRQNVIKTAIKQISLAYSRIYIKDIAKKLYITNETETEYIVAKAIADGA 414
Query 123 IDASIDHD----KQFLQSKASVDLYASNEPQKAFHRRITFCLDLYNDAVKAMQYPGSTER 178
IDA I D +++QS + D+Y ++EPQ F RI +CL+L+N AVKA++YP +
Sbjct 415 IDAVITSDVRDGPRYMQSSETADIYRTSEPQAHFDTRIRYCLELHNQAVKALRYPPKKKI 474
Query 179 QQTAEDDERKRAMQEERARAEDEDLGDD 206
+ R+R QE E D DD
Sbjct 475 AVETIEQAREREQQELEFAKELADEDDD 502
> ECU05g1540
Length=376
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 62/130 (47%), Gaps = 14/130 (10%)
Query 2 LAALKKCIVVELLMGDI----PERPLFNKQNMHAELVPYREIVLAVRSGDLHAFASVLQQ 57
L A K+ I+ LL D P +P L Y ++ AV+ D+ F L+
Sbjct 203 LGAEKRVILCMLLSSDYSIPYPCKP---------SLRIYFKLASAVKRADIKKFEETLES 253
Query 58 HEQRFRLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGKA 117
++ Y + RL QNVI+ G+R IS+ YSRI +DIA LG++S E E + +
Sbjct 254 NKDELMSQGLYFVAKRLSQNVIQEGIRKISVVYSRISYEDIAHILGINSG-EVEYLVKRT 312
Query 118 ILDRVIDASI 127
I +I +
Sbjct 313 IRKGLIKGKV 322
> Hs4502975
Length=403
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 46/198 (23%), Positives = 89/198 (44%), Gaps = 27/198 (13%)
Query 2 LAALKKCIVVEL-LMGDIPERPLFNKQNMHAELVP----YREIVLAVRSGDLHAFASVLQ 56
L + KK I+V L L+G + + P + Q + + P Y E+ + + +++
Sbjct 201 LESYKKYILVSLILLGKVQQLPKYTSQIVGRFIKPLSNAYHELAQVYSTNNPSELRNLVN 260
Query 57 QHEQRFRLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGK 116
+H + F D+ L+ + ++ + ++ ++ ++ + L+D+A ++ L +EAE
Sbjct 261 KHSETFTRDNNMGLVKQCLSSLYKKNIQRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLH 320
Query 117 AILDRVIDASIDHDKQFLQSKASVDLYASNEPQKAFHRRITFCLDLYNDAVKAMQYPGST 176
I D I ASI+ Q V + + P+K YN+ AM +
Sbjct 321 MIEDGEIFASIN------QKDGMVSFH--DNPEK------------YNNP--AMLHNIDQ 358
Query 177 ERQQTAEDDERKRAMQEE 194
E + E DER +AM +E
Sbjct 359 EMLKCIELDERLKAMDQE 376
> At2g19560
Length=244
Score = 33.1 bits (74), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 36 YREIVLAVRSGDLHAFASVLQQHEQRFRLDDTYTLLCRLHQNV 78
Y +IV A+R GDL LQ+HE RF Y +L +L V
Sbjct 194 YTKIVQALRKGDLRLLRHALQEHEDRFLRSGVYLVLEKLELQV 236
> Hs4506221
Length=456
Score = 32.7 bits (73), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 59/123 (47%), Gaps = 20/123 (16%)
Query 15 MGDIPERPLFNKQNMHAELVPYREIV----LAVRSGDLHA-----FASVLQQHEQRFRLD 65
+ +IP+ K EL+ + +V + +R G L + F S ++ E+R++
Sbjct 296 LEEIPKYKDLLKLFTTMELMRWSTLVEDYGMELRKGSLESPATDVFGST-EEGEKRWK-- 352
Query 66 DTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGKAILDRVIDA 125
L V+ +RI++ Y+RI +K +A+ L L S E+EA ++++ I A
Sbjct 353 -------DLKNRVVEHNIRIMAKYYTRITMKRMAQLLDL-SVDESEAFLSNLVVNKTIFA 404
Query 126 SID 128
+D
Sbjct 405 KVD 407
> At3g56170
Length=276
Score = 32.7 bits (73), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 8/100 (8%)
Query 61 RFRLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKL-GLDSAKEAEAVAGK--- 116
+ R YT LH+ +I +G R+IS + K +L G+DS E++ GK
Sbjct 173 KAREKKNYTEADALHKTIIASGYRMISFQNEEVLAKKFRIRLSGIDSP-ESKMPYGKEAH 231
Query 117 -AILDRVIDASIDHDKQFLQSKASVDLYASNEPQKAFHRR 155
+L V + + + Q + V L+AS+ P+K + R
Sbjct 232 DELLKMVEGKCLKWENEARQKR--VGLWASSNPEKPWEWR 269
> Hs22061203
Length=607
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 41/93 (44%), Gaps = 8/93 (8%)
Query 58 HEQRFRLDDTYTLLCRLHQN--VIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAG 115
H + +DDT+ C L ++ L + YS+I+L I + LG SA ++AG
Sbjct 470 HGKLTFVDDTFGFPCLLASGGPLLSVSLHFSAYVYSQIHLAFILRDLGSHSAPSLASLAG 529
Query 116 K------AILDRVIDASIDHDKQFLQSKASVDL 142
++LDR D + +S VDL
Sbjct 530 PRELTVGSLLDREWRQIKTDDFELGKSAGEVDL 562
> Hs18554904
Length=113
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 45/87 (51%), Gaps = 11/87 (12%)
Query 42 AVRSGDLHAFASVLQQHEQRFRLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKK 101
+V S AF S ++ E+R++ L+ V+ +RI++ Y+R+ +K + +
Sbjct 21 SVGSPATDAFGST-EEGEKRWK---------DLNNRVVEHNIRIMAKYYTRMTMKRMVQL 70
Query 102 LGLDSAKEAEAVAGKAILDRVIDASID 128
L L S E+EA ++++ I A +D
Sbjct 71 LDL-SVDESEAFLSNLVVNKTIFAKVD 96
> 7296672
Length=502
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 36/129 (27%), Positives = 62/129 (48%), Gaps = 15/129 (11%)
Query 27 QNMHAELVP-YREIVLAVRSGDLHAFAS-------VLQQHEQRFRLDDTYTLLC--RLHQ 76
+N E VP Y+EI+ S +L F + VL ++E F+ + C L
Sbjct 339 KNKKLEEVPAYKEILRLFMSKELINFDTFNADFGLVLAENEM-FKDSTKHGKKCITELKD 397
Query 77 NVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGKAILDRVIDASIDHDK---QF 133
+I +RII++ YSR++L +++ L L +++ E ++ A D I ID F
Sbjct 398 RLIEHNIRIIAMYYSRLHLARMSELLNLPTSRCEEYLSKLANTD-TIRVKIDRPAGIIYF 456
Query 134 LQSKASVDL 142
Q K++ D+
Sbjct 457 TQKKSASDI 465
> ECU07g0590
Length=389
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 41/89 (46%), Gaps = 12/89 (13%)
Query 47 DLHAFASVLQQHEQRFRLDD------TYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAK 100
DL +++ VL Q+ + D Y L +N+++ I YS + ++ IA
Sbjct 272 DLKSYSDVLAQYSDQIHQDSFIRDHLQYLYDVLLDKNIVK-----IIEPYSVVKIRFIAD 326
Query 101 KLGLDSAKEAEAVAGKAILDRVIDASIDH 129
LG D E K ILD+ +D ++DH
Sbjct 327 VLGFD-VNVIEKKLRKMILDKAVDGTLDH 354
> 7296363
Length=445
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 39/165 (23%), Positives = 72/165 (43%), Gaps = 6/165 (3%)
Query 2 LAALKKCIVVELLM-GDIPERPLFNKQNMHAELVP----YREIVLAVRSGDLHAFASVLQ 56
L A KK ++V L++ G I P N Q + + P Y ++V + ++
Sbjct 238 LEAYKKFLMVSLIVEGKIAYIPK-NTQVIGRFMKPMANHYHDLVNVYANSSSEELRIIIL 296
Query 57 QHEQRFRLDDTYTLLCRLHQNVIRAGLRIISLSYSRIYLKDIAKKLGLDSAKEAEAVAGK 116
++ + F D+ L ++ ++ + ++ ++ ++ + L D+A ++ L SA EAE
Sbjct 297 KYSEAFTRDNNMGLAKQVATSLYKRNIQRLTKTFLTLSLSDVASRVQLASAVEAERYILN 356
Query 117 AILDRVIDASIDHDKQFLQSKASVDLYASNEPQKAFHRRITFCLD 161
I I ASI+ + K + Y S E IT LD
Sbjct 357 MIKSGEIYASINQKDGMVLFKDDPEKYNSPEMFLNVQNNITHVLD 401
Lambda K H
0.321 0.135 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3790221436
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40