bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3930_orf2
Length=278
Score E
Sequences producing significant alignments: (Bits) Value
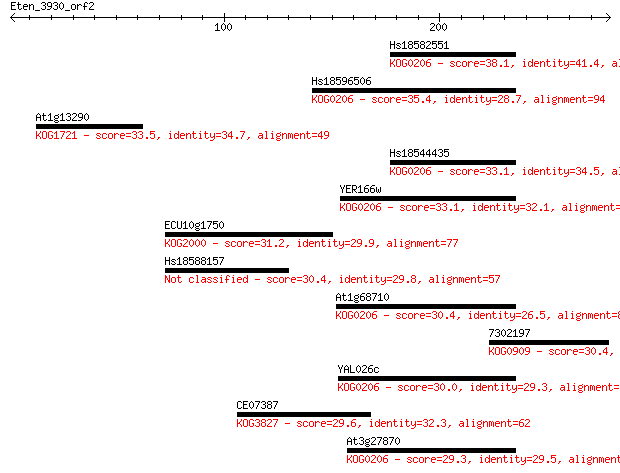
Hs18582551 38.1 0.019
Hs18596506 35.4 0.15
At1g13290 33.5 0.52
Hs18544435 33.1 0.73
YER166w 33.1 0.75
ECU10g1750 31.2 2.5
Hs18588157 30.4 3.7
At1g68710 30.4 4.4
7302197 30.4 4.6
YAL026c 30.0 5.3
CE07387 29.6 8.1
At3g27870 29.3 9.1
> Hs18582551
Length=782
Score = 38.1 bits (87), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 177 LVYAICRADTNV-CAEVRGQLKQQLVACIKASLKPQPIVIAAGSSAEDAAMMREATIGI 234
L ICR+ + V C + K Q+V IK S K PI +A G A D +M+ EA +GI
Sbjct 431 LFLEICRSCSAVLCCRMAPLQKAQIVKLIKFS-KEHPITLAIGDGANDVSMILEAHVGI 488
> Hs18596506
Length=950
Score = 35.4 bits (80), Expect = 0.15, Method: Composition-based stats.
Identities = 27/100 (27%), Positives = 46/100 (46%), Gaps = 11/100 (11%)
Query 141 FEDMWRQSRVHGQGICLLSDAASLAFFFSHKLLQGLLVYA-----ICRADTNV-CAEVRG 194
F+ W + + +G L+ D ++L+ + Y IC T V C +
Sbjct 666 FKKAWTEHQEYG----LIIDGSTLSLILNSSQDSSSNNYKSIFLQICMKCTAVLCCRMAP 721
Query 195 QLKQQLVACIKASLKPQPIVIAAGSSAEDAAMMREATIGI 234
K Q+V +K +LK PI ++ G A D +M+ E+ +GI
Sbjct 722 LQKAQIVRMVK-NLKGSPITLSIGDGANDVSMILESHVGI 760
> At1g13290
Length=302
Score = 33.5 bits (75), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 23/49 (46%), Gaps = 2/49 (4%)
Query 13 RCSAELTYYARKDRRALQSPSASLWACECGTTVSRRRVLKTSIRRRSFG 61
R E T+ R D R + LW C CG+ +R LK + R+FG
Sbjct 189 RKKCEKTFAVRGDWRTHEKNCGKLWFCVCGSDFKHKRSLKDHV--RAFG 235
> Hs18544435
Length=761
Score = 33.1 bits (74), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 177 LVYAICRADTNV-CAEVRGQLKQQLVACIKASLKPQPIVIAAGSSAEDAAMMREATIGI 234
L +CR + V C + K +++ IK S +PI +A G A D +M++EA +GI
Sbjct 363 LFMEVCRNCSAVLCCRMAPLQKAKVIRLIKIS-PEKPITLAVGDGANDVSMIQEAHVGI 420
> YER166w
Length=1571
Score = 33.1 bits (74), Expect = 0.75, Method: Composition-based stats.
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Query 154 GICLLSDAASLAFFFSHKLLQGLLVYAICRADTNVCAEVRGQLKQQLVACIKASLKPQPI 213
I + DA LA + + LL+ CRA +C V K +V +K SL +
Sbjct 1069 AIVIDGDALKLALYGEDIRRKFLLLCKNCRA--VLCCRVSPSQKAAVVKLVKDSLDVMTL 1126
Query 214 VIAAGSSAEDAAMMREATIGI 234
I GS+ D AM++ A +GI
Sbjct 1127 AIGDGSN--DVAMIQSADVGI 1145
> ECU10g1750
Length=618
Score = 31.2 bits (69), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 39/85 (45%), Gaps = 9/85 (10%)
Query 73 DRSCSRTRGYFGRDRDATLYQREILNKDLRPRRRVLRHRIETKEGQNVSLSSLPLRRKLR 132
D ++ G G D + Q ILN DL ++ H+I K+ ++V L+S +R ++
Sbjct 321 DVESAKDAGLLGEDAEPNSKQINILNPDLEENIDIMLHKI-NKKIEHVFLNSPSIRNQIE 379
Query 133 -------IGRC-FVQVFEDMWRQSR 149
GRC F++ + SR
Sbjct 380 GIKNTFLFGRCDFIETLFLYLKDSR 404
> Hs18588157
Length=459
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 35/62 (56%), Gaps = 6/62 (9%)
Query 73 DRSCSRTRGYFG-----RDRDATLYQREILNKDLRPRRRVLRHRIETKEGQNVSLSSLPL 127
D SCS+++GY G +D T + ++ +++ PR +VL R+ T E ++ +++
Sbjct 396 DGSCSKSKGYGGPGNQTKDSSKTTIVKTVV-EEIDPRGKVLSSRVHTVEEKSTKVNNKNE 454
Query 128 RR 129
+R
Sbjct 455 QR 456
> At1g68710
Length=1200
Score = 30.4 bits (67), Expect = 4.4, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 32/83 (38%), Gaps = 3/83 (3%)
Query 152 GQGICLLSDAASLAFFFSHKLLQGLLVYAICRADTNVCAEVRGQLKQQLVACIKASLKPQ 211
G L+ D SLA+ + L A+ A C R KQ+ +
Sbjct 806 GNAFALIIDGKSLAYALDDDIKHIFLELAVSCASVICC---RSSPKQKALVTRLVKSGNG 862
Query 212 PIVIAAGSSAEDAAMMREATIGI 234
+A G A D M++EA IG+
Sbjct 863 KTTLAIGDGANDVGMLQEADIGV 885
> 7302197
Length=591
Score = 30.4 bits (67), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 24/55 (43%), Gaps = 0/55 (0%)
Query 223 DAAMMREATIGILMLQALTPAEEATYVAEGPAEPMADPVTSSENAAKPPVARHLV 277
DA M E + + + L P EE T +A + D + S E K P R L+
Sbjct 171 DAVMQYEDNLLLATGRTLIPVEELTEMASEKLIDIQDQIASGERQEKEPCVRDLL 225
> YAL026c
Length=1355
Score = 30.0 bits (66), Expect = 5.3, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 40/83 (48%), Gaps = 5/83 (6%)
Query 153 QGICLLSDAASLAFFFSHKLLQGLLVYA-ICRADTNVCAEVRGQLKQQLVACIKASLKPQ 211
+ + L+ D SL F +L LL A +C+A +C V K +V +K
Sbjct 891 KSLALVIDGKSLGFALEPELEDYLLTVAKLCKA--VICCRVSPLQKALVVKMVKRKSSSL 948
Query 212 PIVIAAGSSAEDAAMMREATIGI 234
+ IA+G A D +M++ A +G+
Sbjct 949 LLAIASG--ANDVSMIQAAHVGV 969
> CE07387
Length=514
Score = 29.6 bits (65), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Query 106 RVLRHRIETKEGQ-NVSLSSLPLRRKLRIGRCFVQVFEDMWRQSRVHGQGICLLSDAASL 164
R +R+R+ K+G N+SL ++P +R+ F V E WR ++ LS S
Sbjct 77 RRIRNRLVQKQGLCNISLKNVPKQRRKYFSDIFTTVIEMKWRWCLLYFS----LSFMISW 132
Query 165 AFF 167
+FF
Sbjct 133 SFF 135
> At3g27870
Length=1174
Score = 29.3 bits (64), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 35/78 (44%), Gaps = 3/78 (3%)
Query 157 LLSDAASLAFFFSHKLLQGLLVYAICRADTNVCAEVRGQLKQQLVACIKASLKPQPIVIA 216
L+ D SL + KL + L AI R ++ +C R KQ+ + +A
Sbjct 791 LVIDGKSLTYALDSKLEKEFLELAI-RCNSVICC--RSSPKQKALVTRLVKNGTGRTTLA 847
Query 217 AGSSAEDAAMMREATIGI 234
G A D M++EA IG+
Sbjct 848 IGDGANDVGMLQEADIGV 865
Lambda K H
0.323 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6056485866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40