bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3821_orf1
Length=211
Score E
Sequences producing significant alignments: (Bits) Value
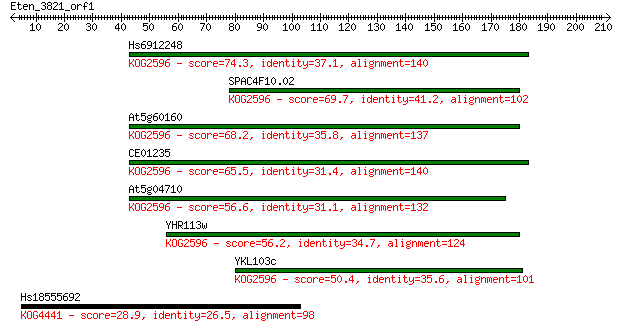
Hs6912248 74.3 2e-13
SPAC4F10.02 69.7 4e-12
At5g60160 68.2 1e-11
CE01235 65.5 8e-11
At5g04710 56.6 3e-08
YHR113w 56.2 4e-08
YKL103c 50.4 3e-06
Hs18555692 28.9 8.7
> Hs6912248
Length=475
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 75/146 (51%), Gaps = 16/146 (10%)
Query 43 EQHLQPVLCSVIAEHLNQGNG-----SSIHEGESEVQRLPPALERLVMQEIGMPGATLLD 97
E HL P+L + I E L +G +++ E V L L+ +G+ +++
Sbjct 184 EMHLVPILATAIQEELEKGTPEPGPLNAVDERHHSV------LMSLLCAHLGLSPKDIVE 237
Query 98 WDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCG-ETEIL 156
+LCL D P L G + EFI +PRLDNL S F A +AL+++ G E +
Sbjct 238 MELCLADTQPAVLGGAYDEFIFAPRLDNLHSCFCALQALIDSCA----GPGSLATEPHVR 293
Query 157 MAVAFDHEEVGSESLAGANSSLLELL 182
M +D+EEVGSES GA S L EL+
Sbjct 294 MVTLYDNEEVGSESAQGAQSLLTELV 319
> SPAC4F10.02
Length=467
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 42/108 (38%), Positives = 59/108 (54%), Gaps = 10/108 (9%)
Query 78 PALERLVMQEIG------MPGATLLDWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFA 131
P L L+ EI + + ++D++L L DA RL GIH EF+ SPRLDNL TF
Sbjct 204 PVLLSLLANEISKSLETTIDPSKIVDFELILGDAEKARLGGIHEEFVFSPRLDNLGMTFC 263
Query 132 AFEALVETSIRQRRGEQDCGETEILMAVAFDHEEVGSESLAGANSSLL 179
A +AL ++ + C + + +FDHEE+GS S GA S+ L
Sbjct 264 ASQALTKSLENNSLDNESC----VRVVPSFDHEEIGSVSAQGAESTFL 307
> At5g60160
Length=477
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 68/144 (47%), Gaps = 10/144 (6%)
Query 43 EQHLQPVLCSVIAEHLNQGNGSSIHEGE-------SEVQRLPPALERLVMQEIGMPGATL 95
+ HL PVL + I LN+ S E S + P L ++ +G +
Sbjct 176 QTHLVPVLATAIKAELNKTPAESGEHDEGKKCAETSSKSKHHPLLMEIIANALGCKPEEI 235
Query 96 LDWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEI 155
D++L D P LAG EFI S RLDNL +F + +AL++ + E + G I
Sbjct 236 CDFELQACDTQPSILAGAAKEFIFSGRLDNLCMSFCSLKALIDATSSGSDLEDESG---I 292
Query 156 LMAVAFDHEEVGSESLAGANSSLL 179
M FDHEEVGS S GA S ++
Sbjct 293 RMVALFDHEEVGSNSAQGAGSPVM 316
> CE01235
Length=470
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 68/143 (47%), Gaps = 7/143 (4%)
Query 43 EQHLQPVLCSVIAEHLN---QGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWD 99
E L+P+L + A +N + + + + P L+ +E G ++D D
Sbjct 179 ETELRPILETFAAAGINAPQKPESTGFADPRNITNNHHPQFLGLIAKEAGCQPEDIVDLD 238
Query 100 LCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMAV 159
L L D + G+ EFI RLDN T+ A L+E+ GE + +I +A
Sbjct 239 LYLYDTNKAAIVGMEDEFISGARLDNQVGTYTAISGLLES----LTGESFKNDPQIRIAA 294
Query 160 AFDHEEVGSESLAGANSSLLELL 182
FD+EEVGS+S GA+SS E +
Sbjct 295 CFDNEEVGSDSAMGASSSFTEFV 317
> At5g04710
Length=526
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 67/132 (50%), Gaps = 3/132 (2%)
Query 43 EQHLQPVLCSVIAEHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWDLCL 102
+ +L+ L ++A ++ + S + S P L +++ ++ ++ +L +
Sbjct 229 KPNLETQLVPLLATKSDESSAESKDKNVSSKDAHHPLLMQILSDDLDCKVEDIVSLELNI 288
Query 103 MDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVETSIRQRRGEQDCGETEILMAVAFD 162
D P L G + EFI S RLDNLAS+F A AL+++ E E +I M FD
Sbjct 289 CDTQPSCLGGANNEFIFSGRLDNLASSFCALRALIDSC---ESSENLSTEHDIRMIALFD 345
Query 163 HEEVGSESLAGA 174
+EEVGS+S GA
Sbjct 346 NEEVGSDSCQGA 357
> YHR113w
Length=490
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 64/128 (50%), Gaps = 15/128 (11%)
Query 56 EHLNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPG-ATLLDWDLCLMDATPGRLAGIH 114
+ +N G +SI ++ VQR L L+ +E+ + + D++L L D L G +
Sbjct 207 KEINNGEFTSI---KTIVQRHHAELLGLIAKELAIDTIEDIEDFELILYDHNASTLGGFN 263
Query 115 LEFIESPRLDNLASTFAAFEALV---ETSIRQRRGEQDCGETEILMAVAFDHEEVGSESL 171
EF+ S RLDNL S F + L +T I + G I + FDHEE+GS S
Sbjct 264 DEFVFSGRLDNLTSCFTSMHGLTLAADTEIDRESG--------IRLMACFDHEEIGSSSA 315
Query 172 AGANSSLL 179
GA+S+ L
Sbjct 316 QGADSNFL 323
> YKL103c
Length=514
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 51/101 (50%), Gaps = 4/101 (3%)
Query 80 LERLVMQEIGMPGATLLDWDLCLMDATPGRLAGIHLEFIESPRLDNLASTFAAFEALVET 139
L R V + G+ + L+ DL L D G + GI F+ +PRLD+ +FAA AL+
Sbjct 259 LLRYVAKLAGVEVSELIQMDLDLFDVQKGTIGGIGKHFLFAPRLDDRLCSFAAMIALICY 318
Query 140 SIRQRRGEQDCGETEILMAVAFDHEEVGSESLAGANSSLLE 180
+ E D T L +D+EE+GS + GA LLE
Sbjct 319 AKDVNTEESDLFSTVTL----YDNEEIGSLTRQGAKGGLLE 355
> Hs18555692
Length=472
Score = 28.9 bits (63), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 26/105 (24%), Positives = 42/105 (40%), Gaps = 7/105 (6%)
Query 5 LLAHSKMNSCCIEVSLHKY-------SSSCPFYLQKQSGEAVFLQEQHLQPVLCSVIAEH 57
+L H K C + ++ ++ P++ + E E HL+ V V+A+
Sbjct 26 MLDHGKFLDCVVRAGEREFPCHRLVLAACSPYFRARFLAEPERAGELHLEEVSPDVVAQV 85
Query 58 LNQGNGSSIHEGESEVQRLPPALERLVMQEIGMPGATLLDWDLCL 102
L+ S I E+ VQ L A R + I + L LCL
Sbjct 86 LHYLYTSEIALDEASVQDLFAAAHRFQIPSIFTICVSFLQKRLCL 130
Lambda K H
0.319 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3825978242
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40