bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3809_orf2
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
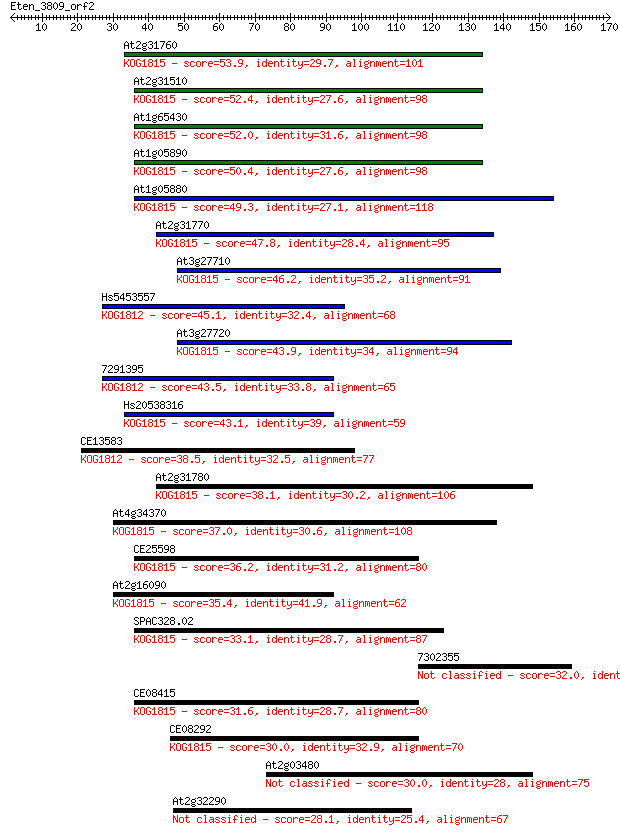
At2g31760 53.9 2e-07
At2g31510 52.4 4e-07
At1g65430 52.0 6e-07
At1g05890 50.4 2e-06
At1g05880 49.3 3e-06
At2g31770 47.8 1e-05
At3g27710 46.2 3e-05
Hs5453557 45.1 7e-05
At3g27720 43.9 2e-04
7291395 43.5 2e-04
Hs20538316 43.1 2e-04
CE13583 38.5 0.006
At2g31780 38.1 0.008
At4g34370 37.0 0.018
CE25598 36.2 0.032
At2g16090 35.4 0.055
SPAC328.02 33.1 0.29
7302355 32.0 0.56
CE08415 31.6 0.88
CE08292 30.0 2.1
At2g03480 30.0 2.2
At2g32290 28.1 9.2
> At2g31760
Length=514
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 54/103 (52%), Gaps = 4/103 (3%)
Query 33 DWDFLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLET 92
D F+ Q++E RR+LK +Y + ++ + ++PLFE+ QG+ E L+ L E
Sbjct 382 DLQFIVDAWLQIIECRRVLKWTYAYGYYLD--NLAKRPLFEYLQGEAETGLERLHHCAEN 439
Query 93 FDPKAYV-DRDPADFF-PFKRQLLDLTAVVRGFFAKICDVFED 133
+ ++ DP+D F F+ +L LT V + +F + E+
Sbjct 440 ELKQFFIKSEDPSDTFNAFRMKLTGLTKVTKTYFDNLVKALEN 482
> At2g31510
Length=565
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 53/99 (53%), Gaps = 1/99 (1%)
Query 36 FLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDP 95
F+ + Q++E RR+LK +Y + ++ E ++ FE+ QG+ E L+ L + +E
Sbjct 358 FILEAWLQIIECRRVLKWTYAYGYYLPEHEHAKRQFFEYLQGEAESGLERLHQCVEKDLV 417
Query 96 KAYVDRDPA-DFFPFKRQLLDLTAVVRGFFAKICDVFED 133
+ + P+ DF F+ +L LT+V + +F + E+
Sbjct 418 QFLIAEGPSKDFNDFRTKLAGLTSVTKNYFENLVKALEN 456
> At1g65430
Length=575
Score = 52.0 bits (123), Expect = 6e-07, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 55/100 (55%), Gaps = 3/100 (3%)
Query 36 FLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDP 95
F+ + Q+VE RR+LK +Y + F+ E ++ FE+ QG+ E L+ L + E +
Sbjct 415 FIIEAWLQIVECRRVLKWTYAYGFYIPDQEHGKRVFFEYLQGEAESGLERLHQCAEK-EL 473
Query 96 KAYVD-RDPA-DFFPFKRQLLDLTAVVRGFFAKICDVFED 133
Y+D + P+ DF F+ +L LT+V + +F + E+
Sbjct 474 LPYLDAKGPSEDFNEFRTKLAGLTSVTKNYFENLVRALEN 513
> At1g05890
Length=545
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/99 (27%), Positives = 54/99 (54%), Gaps = 3/99 (3%)
Query 36 FLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDP 95
F+ + Q++E RR+LK +Y + ++ + + +KP FE+ QG+ E L+ L + +E
Sbjct 404 FIAEAWLQIIECRRVLKWTYAYGYYLQ--DHAKKPFFEYLQGEAESGLERLHKCVEKDIE 461
Query 96 KAYVDRDPA-DFFPFKRQLLDLTAVVRGFFAKICDVFED 133
+ P+ +F F+ +L LT++ + FF + E+
Sbjct 462 VFELAEGPSEEFNHFRTKLTGLTSITKTFFENLVKALEN 500
> At1g05880
Length=426
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 64/120 (53%), Gaps = 5/120 (4%)
Query 36 FLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLET-FD 94
F+ + G Q++E RR+L+ +Y + ++ E ++ L + Q +L++ ++ L+ LET
Sbjct 310 FILEAGLQIIECRRVLEWTYVYGYYLREDEVGKQNLLKDTQERLKKFVENLKHCLETNLQ 369
Query 95 PKAYVDRDPADFFPFKRQLLDLTAVVRGFFAKICDVFEDEFVAPPVPVNERE-TGWGKKQ 153
P Y + DF F+ +L +LT++ R + + E+ + V+E E +G G+ Q
Sbjct 370 PFRYEEEPSKDFNAFRIKLTELTSLTRNHYENVVKDVENGLAS---VVSEGEASGSGRNQ 426
> At2g31770
Length=543
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 51/97 (52%), Gaps = 4/97 (4%)
Query 42 FQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDPKAYV-D 100
Q++E RR+LK +Y + ++ + + ++ FE+ QG+ E L+ L E + ++
Sbjct 401 LQIIECRRVLKWTYAYGYYLQ--DLPKRKFFEYLQGEAESGLERLHHCAENELKQFFIKS 458
Query 101 RDPADFF-PFKRQLLDLTAVVRGFFAKICDVFEDEFV 136
DP+D F F+ +L LT V + +F + E+ V
Sbjct 459 EDPSDTFNAFRMKLTGLTTVTKTYFENLVKALENGLV 495
> At3g27710
Length=537
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 32/102 (31%), Positives = 49/102 (48%), Gaps = 15/102 (14%)
Query 48 RRMLKGSYPFAFF-----------AEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDPK 96
RR+L SYPFAF+ +E +K LFE Q QLE +++ L + LE +
Sbjct 405 RRILSQSYPFAFYMFGEELFKDEMSEKEREIKKNLFEDQQQQLEGNVEKLSKILE----E 460
Query 97 AYVDRDPADFFPFKRQLLDLTAVVRGFFAKICDVFEDEFVAP 138
+ + D R L +LTAVV ++ + E+E + P
Sbjct 461 PFDEYDHEKVVEMMRHLTNLTAVVDNLCKEMYECIENELLGP 502
> Hs5453557
Length=493
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Query 27 GFGEDFDWDFLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLL 86
G DW +L+ + + R L+ +YP+A++ E +K LFE+ Q QLE ++ L
Sbjct 400 NLGTWIDWQYLQNAAKLLAKCRYTLQYTYPYAYYME--SGPRKKLFEYQQAQLEAEIENL 457
Query 87 QEKLETFD 94
K+E D
Sbjct 458 SWKVERAD 465
> At3g27720
Length=493
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 50/106 (47%), Gaps = 17/106 (16%)
Query 48 RRMLKGSYPFAFF-----------AEFPESRQKPLFEFHQGQLERSLDLLQEKLET-FDP 95
RR+L SYPF F+ ++ + +K LFE Q QLE +++ L + LE FD
Sbjct 365 RRILSYSYPFVFYMFGKELFKDDMSDEERNIKKNLFEDQQQQLEGNVERLSKILEEPFD- 423
Query 96 KAYVDRDPADFFPFKRQLLDLTAVVRGFFAKICDVFEDEFVAPPVP 141
+ D R L +LTAVV ++ + E+E + P +
Sbjct 424 ----EYDHEKVVEMMRHLTNLTAVVDNLCKEMYECIENELLGPLIS 465
> 7291395
Length=509
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query 27 GFGEDFDWDFLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLL 86
G G DW +L + + R L+ +YP+A++ E +K LFE+ Q QLE ++ L
Sbjct 414 GSGTWIDWQYLFNAAALLAKCRYTLQYTYPYAYYME--AGSRKNLFEYQQAQLEAEIENL 471
Query 87 QEKLE 91
K+E
Sbjct 472 SWKIE 476
> Hs20538316
Length=1089
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 33 DWDFLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLE 91
D F+E +++ RR+LK SYP+ FF E P+S +K +FE Q LE + L +K+
Sbjct 642 DTTFIEDAVHVLLKTRRILKCSYPYGFFLE-PKSTKKEIFELMQTDLEMVTEDLAQKVN 699
> CE13583
Length=488
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 40/83 (48%), Gaps = 8/83 (9%)
Query 21 DPRYSLGFGEDFDWDFLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPL------FEF 74
D + + G DW +L K + + R L+ +YPFA+F + +K L FE+
Sbjct 384 DDKVNEHNGTWIDWQYLHKSVSLLTKCRYTLQYTYPFAYF--LSATPRKNLVCFLGKFEY 441
Query 75 HQGQLERSLDLLQEKLETFDPKA 97
Q QLE+ ++ L +E D A
Sbjct 442 QQAQLEKEVEELAWAVERADGTA 464
> At2g31780
Length=542
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 32/118 (27%), Positives = 55/118 (46%), Gaps = 19/118 (16%)
Query 42 FQVVEFRRMLKGSYPFAFFAEFPESRQKPLF-----------EFHQGQLERSLDLLQEKL 90
Q++E RR+LK +Y + ++ + R K +F E G LER +E+L
Sbjct 412 LQIIECRRVLKWTYAYGYYI-LSQERNKRVFARTFSLSCCSAEAENG-LERLHHCAEEEL 469
Query 91 ETFDPKAYVDRDPA-DFFPFKRQLLDLTAVVRGFFAKICDVFEDEFVAPPVPVNERET 147
+ F K DP+ +F + +L+DLT + +F + E+ V V NE ++
Sbjct 470 KQFIGKI---EDPSKNFGELRAKLIDLTKATKTYFENLVKALENGLV--DVAYNESQS 522
> At4g34370
Length=594
Score = 37.0 bits (84), Expect = 0.018, Method: Composition-based stats.
Identities = 33/119 (27%), Positives = 55/119 (46%), Gaps = 17/119 (14%)
Query 30 EDFDWDFLEKGGFQVVEFRRMLKGSYPFAF--FAE-------FPESRQ--KPLFEFHQGQ 78
+DF W + G ++ RR+L SY FA+ F E PE R+ K LFE Q Q
Sbjct 389 KDFSW--VTNGLDRLFRSRRVLSYSYAFAYYMFGEEMFKDEMTPEEREIKKNLFEDQQQQ 446
Query 79 LERSLDLLQEKLETFDPKAYVDRDPADFFPFKRQLLDLTAVVRGFFAKICDVFEDEFVA 137
LE ++ EKL F + + + + Q+++L+ V K+ + E++ +
Sbjct 447 LESNV----EKLSQFLEEPFDEFSNDKVMAIRIQIINLSVAVDTLCKKMYECIENDLLG 501
> CE25598
Length=485
Score = 36.2 bits (82), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 37/80 (46%), Gaps = 9/80 (11%)
Query 36 FLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDP 95
FL + + E RR LK +Y FA++ E + LFE +Q LE + + L LE
Sbjct 386 FLRRAVDALCECRRTLKYAYAFAYYLE--ANNMTTLFETNQSDLELATEQLSGMLEG--- 440
Query 96 KAYVDRDPADFFPFKRQLLD 115
D + D KR++ D
Sbjct 441 ----DLEDNDLAELKRKVQD 456
> At2g16090
Length=518
Score = 35.4 bits (80), Expect = 0.055, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 37/74 (50%), Gaps = 15/74 (20%)
Query 30 EDFDWDFLEKGGFQVVEFRRMLKGSYPFAFF------------AEFPESRQKPLFEFHQG 77
+DF W G ++ RR+L SYPFAF+ +E E +Q LFE Q
Sbjct 361 KDFSW--ATNGLHRLFRSRRVLSYSYPFAFYMFGDELFKDEMSSEEREIKQN-LFEDQQQ 417
Query 78 QLERSLDLLQEKLE 91
QLE +++ L + LE
Sbjct 418 QLEANVEKLSKFLE 431
> SPAC328.02
Length=504
Score = 33.1 bits (74), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 40/87 (45%), Gaps = 9/87 (10%)
Query 36 FLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDP 95
FL+ + + R+ LK +Y FA++ + Q +FE +Q R L+L E L
Sbjct 403 FLKNAVDILFQCRQTLKWTYAFAYY--LARNNQTEIFEDNQ----RDLELAVENLSELCE 456
Query 96 KAYVDRDPADFFPFKRQLLDLTAVVRG 122
+ D FK+++LD T VR
Sbjct 457 RPCQD---CSLSVFKQRVLDKTVYVRS 480
> 7302355
Length=1469
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 20/43 (46%), Gaps = 0/43 (0%)
Query 116 LTAVVRGFFAKICDVFEDEFVAPPVPVNERETGWGKKQQPPAL 158
T + F A+I + F+ APP P N +E K Q P L
Sbjct 1298 TTTAQKAFAAEIMNAFQKVTTAPPKPSNAKEAAKPKSQAAPKL 1340
> CE08415
Length=491
Score = 31.6 bits (70), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 13/82 (15%)
Query 36 FLEKGGFQVVEFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDP 95
FL K + E RR L +Y FAF+ + +FE +Q LE +ET
Sbjct 392 FLRKAVDILSECRRTLMYTYAFAFY--LKKDNNSIIFESNQANLE---------METEQL 440
Query 96 KAYVDRD--PADFFPFKRQLLD 115
+++RD D K+++ D
Sbjct 441 SGFLERDLEDEDLVTLKQKVQD 462
> CE08292
Length=115
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 33/70 (47%), Gaps = 9/70 (12%)
Query 46 EFRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDPKAYVDRDPAD 105
E RR LK +Y FA++ E + LFE +Q LE + + L LE D + D
Sbjct 18 ECRRTLKYAYAFAYYLE--ANNLTTLFETNQSDLELATEQLSGMLEG-------DLEDMD 68
Query 106 FFPFKRQLLD 115
KR++ D
Sbjct 69 LAELKRKVQD 78
> At2g03480
Length=380
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 32/75 (42%), Gaps = 0/75 (0%)
Query 73 EFHQGQLERSLDLLQEKLETFDPKAYVDRDPADFFPFKRQLLDLTAVVRGFFAKICDVFE 132
+ + L+ LL + + PK D DP F R ++D+ A A + D +
Sbjct 197 QIWRSALKNYWSLLTPLIFSDHPKRPGDEDPLPPFNMIRNVMDMHARFGNLNAALLDEGK 256
Query 133 DEFVAPPVPVNERET 147
+V VPVN R T
Sbjct 257 SAWVMNVVPVNARNT 271
> At2g32290
Length=505
Score = 28.1 bits (61), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 29/67 (43%), Gaps = 0/67 (0%)
Query 47 FRRMLKGSYPFAFFAEFPESRQKPLFEFHQGQLERSLDLLQEKLETFDPKAYVDRDPADF 106
F+ LK + + PE L + + + SL++ E+ E FDP ++D
Sbjct 427 FKMFLKRMHANQEYCSEPERYNHELLPLERSRNDESLEMFMEETEPFDPFPWLDETDMSI 486
Query 107 FPFKRQL 113
PF+ L
Sbjct 487 RPFESVL 493
Lambda K H
0.323 0.144 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2454498488
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40