bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3797_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
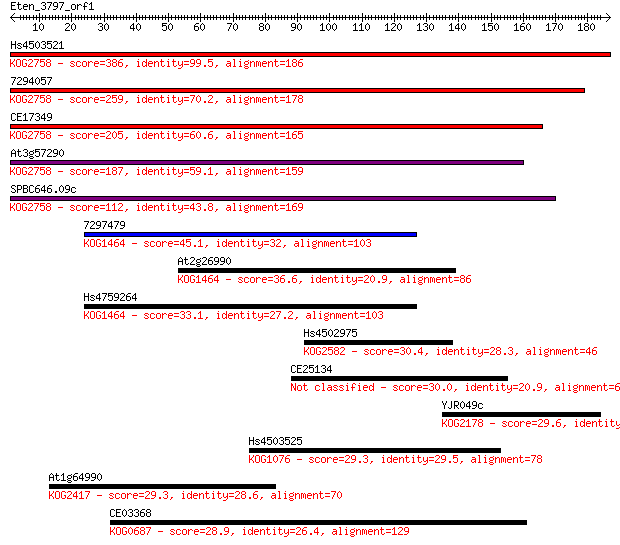
Hs4503521 386 2e-107
7294057 259 2e-69
CE17349 205 4e-53
At3g57290 187 1e-47
SPBC646.09c 112 3e-25
7297479 45.1 7e-05
At2g26990 36.6 0.031
Hs4759264 33.1 0.32
Hs4502975 30.4 2.1
CE25134 30.0 2.5
YJR049c 29.6 4.0
Hs4503525 29.3 4.7
At1g64990 29.3 5.1
CE03368 28.9 6.8
> Hs4503521
Length=445
Score = 386 bits (991), Expect = 2e-107, Method: Compositional matrix adjust.
Identities = 185/186 (99%), Positives = 185/186 (99%), Gaps = 0/186 (0%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL
Sbjct 260 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 319
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR
Sbjct 320 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 379
Query 121 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAA 180
LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWA
Sbjct 380 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAT 439
Query 181 QDSGFY 186
QDSGFY
Sbjct 440 QDSGFY 445
> 7294057
Length=435
Score = 259 bits (663), Expect = 2e-69, Method: Compositional matrix adjust.
Identities = 125/179 (69%), Positives = 148/179 (82%), Gaps = 4/179 (2%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AV+ N R RR LKDL+KVIQQESYTY+DPITEF+ECLYVNFDF+GA+ KL EC++V+
Sbjct 257 AVVIN---RTRRNALKDLIKVIQQESYTYRDPITEFLECLYVNFDFEGARLKLHECQTVI 313
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
+NDFF+VACL +F+E+ARL IFETFCRIHQCI+I+MLADKLNM P EAE WIVNLIRNAR
Sbjct 314 LNDFFIVACLNEFVEDARLMIFETFCRIHQCITISMLADKLNMKPNEAECWIVNLIRNAR 373
Query 121 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSE-APNW 178
L+AKIDSKLGHVVMG +SPYQQ++EK SLS RS+ LA IE+K Q E A +W
Sbjct 374 LNAKIDSKLGHVVMGTQPLSPYQQLVEKIDSLSMRSEHLAGLIERKSKQKQNQESADSW 432
> CE17349
Length=432
Score = 205 bits (521), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 100/166 (60%), Positives = 131/166 (78%), Gaps = 3/166 (1%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AV+T+K +++ LKDLVKVI E ++YKDP+T+F+ CLY+ +DFD AQ+ L++CE VL
Sbjct 260 AVVTSKS--RQKNSLKDLVKVIDIERHSYKDPVTDFLTCLYIKYDFDEAQEMLQKCEEVL 317
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
NDFFL A L DF E+ARL IFE FCRIHQCI+I MLA +LNM+ EEAERWIV+LIR R
Sbjct 318 SNDFFLTAVLGDFRESARLLIFEMFCRIHQCITIEMLARRLNMSQEEAERWIVDLIRTYR 377
Query 121 LD-AKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEK 165
++ AKIDSKLG VVMG +VS ++QV+E TK L+ R+Q +A+ +EK
Sbjct 378 IEGAKIDSKLGQVVMGVKSVSIHEQVMENTKRLTLRAQQIALQLEK 423
> At3g57290
Length=418
Score = 187 bits (475), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 94/170 (55%), Positives = 123/170 (72%), Gaps = 14/170 (8%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
A I NK +RR LK+ +KVIQQE Y+YKDPI EF+ C++VN+DFDGAQKK++ECE V+
Sbjct 238 AFIVNK---RRRPQLKEFIKVIQQEHYSYKDPIIEFLACVFVNYDFDGAQKKMKECEEVI 294
Query 61 VNDFFLVACLED-----------FIENARLFIFETFCRIHQCISINMLADKLNMTPEEAE 109
VND FL +ED F+ENARLF+FET+C+IHQ I + +LA+KLN+ EEAE
Sbjct 295 VNDPFLGKRVEDGNFSTVPLRDEFLENARLFVFETYCKIHQRIDMGVLAEKLNLNYEEAE 354
Query 110 RWIVNLIRNARLDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQML 159
RWIVNLIR ++LDAKIDS+ G V+M + ++Q+I TK LS R+ L
Sbjct 355 RWIVNLIRTSKLDAKIDSESGTVIMEPTQPNVHEQLINHTKGLSGRTYKL 404
> SPBC646.09c
Length=501
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 74/229 (32%), Positives = 112/229 (48%), Gaps = 64/229 (27%)
Query 1 AVITNKD-----VRKRRQV----LKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQK 51
AV+TN++ R RQ ++DLV++I QE+Y Y DP+T F+ LY DF+ AQ
Sbjct 271 AVVTNQNNANQKPRNPRQSYQRRMRDLVRIISQENYEYSDPVTSFISALYTEVDFEKAQH 330
Query 52 KLRECESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERW 111
LRECE VL DFFLV+ + F+E AR + E +CRIH IS+++LA+KL M +
Sbjct 331 CLRECEEVLKTDFFLVSLCDHFLEGARKLLAEAYCRIHSVISVDVLANKLEMDSAQ---- 386
Query 112 IVNLIRN----------------ARLDAKIDSKLGHVV---------------------- 133
++ L+ N + D I+S +VV
Sbjct 387 LIQLVENRNNPSVAAASNVAADQSTEDESIESTSTNVVADDLITEAETATEAEEPEPEVQ 446
Query 134 -------------MGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQ 169
+ + S +QQ+I++TKSLSF SQ L ++ K +++
Sbjct 447 FGFKAKLDGESIIIEHPTYSAFQQIIDRTKSLSFESQNLEQSLAKSISE 495
> 7297479
Length=444
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 55/108 (50%), Gaps = 6/108 (5%)
Query 24 QESYTYK-DP----ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENAR 78
QE+ YK DP +T V Y N D + + LR+ S ++ D F+ +ED + N R
Sbjct 298 QEAKPYKNDPEILAMTNLVNS-YQNNDINEFETILRQHRSNIMADQFIREHIEDLLRNIR 356
Query 79 LFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKID 126
+ R ++ I+I +A+ LN+ P E E +V+ I + + +ID
Sbjct 357 TQVLIKLIRPYKNIAIPFIANALNIEPAEVESLLVSCILDDTIKGRID 404
> At2g26990
Length=433
Score = 36.6 bits (83), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 18/86 (20%), Positives = 44/86 (51%), Gaps = 0/86 (0%)
Query 53 LRECESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWI 112
L+ +++D F+ +ED ++ R + + + I I ++ +LN+ + +
Sbjct 320 LKSNRRTIMDDPFIRNYMEDLLKKVRTQVLLKLIKPYTKIGIPFISKELNVPETDVTELL 379
Query 113 VNLIRNARLDAKIDSKLGHVVMGNNA 138
V+LI ++R+D ID +++ G++
Sbjct 380 VSLILDSRIDGHIDEMNRYLLRGDSG 405
> Hs4759264
Length=443
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 6/108 (5%)
Query 24 QESYTYK-DP----ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENAR 78
QE+ YK DP +T V Y N D +K L+ S +++D F+ +E+ + N R
Sbjct 297 QEAKPYKNDPEILAMTNLVSA-YQNNDITEFEKILKTNHSNIMDDPFIREHIEELLRNIR 355
Query 79 LFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKID 126
+ + + I I ++ +LN+ + E +V I + + +ID
Sbjct 356 TQVLIKLIKPYTRIHIPFISKELNIDVADVESLLVQCILDNTIHGRID 403
> Hs4502975
Length=403
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 92 ISINMLADKLNMT-PEEAERWIVNLIRNARLDAKIDSKLGHVVMGNN 137
+S+ +A ++ ++ P+EAE++++++I + + A I+ K G V +N
Sbjct 297 LSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDN 343
> CE25134
Length=549
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/67 (20%), Positives = 35/67 (52%), Gaps = 2/67 (2%)
Query 88 IHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVMGNNAVSPYQQVIE 147
I +C+++ + +++ P + + +++ R D + KL +++G A +PY++ E
Sbjct 124 IQKCLAVGYYSRAIDLDPNQGRAF--HVLAGLRADLNVAQKLRLMILGQLADAPYKKGTE 181
Query 148 KTKSLSF 154
+ L F
Sbjct 182 LLEYLKF 188
> YJR049c
Length=530
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 26/53 (49%), Gaps = 4/53 (7%)
Query 135 GNNAVSPYQQVIEKTK----SLSFRSQMLAMNIEKKLNQNSRSEAPNWAAQDS 183
G + V P I T +LSFR +L +I K+ + +S AP WAA D
Sbjct 350 GGSLVCPTVNAIALTPICPHALSFRPIILPESINLKVKVSMKSRAPAWAAFDG 402
> Hs4503525
Length=913
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 5/82 (6%)
Query 75 ENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVM 134
E+ R ++F T+ ++ IS+ L+D + I +I N L A +D VVM
Sbjct 786 ESLRTYLF-TYSSVYDSISMETLSDMFELDLPTVHSIISKMIINEELMASLDQPTQTVVM 844
Query 135 GNNAVSPYQ----QVIEKTKSL 152
+ Q Q+ EK SL
Sbjct 845 HRTEPTAQQNLALQLAEKLGSL 866
> At1g64990
Length=465
Score = 29.3 bits (64), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 1/70 (1%)
Query 13 QVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLED 72
++LK L V+ +E+ T KDP+T + FD L + S+L +V +
Sbjct 306 KMLKSLQSVVFKEAGT-KDPVTTMISIFLRLFDIGVDAALLSQYISLLFIGMLIVISVRG 364
Query 73 FIENARLFIF 82
F+ N F F
Sbjct 365 FLTNLMKFFF 374
> CE03368
Length=410
Score = 28.9 bits (63), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 34/133 (25%), Positives = 59/133 (44%), Gaps = 6/133 (4%)
Query 32 PITEFVECLYVNFDFDGAQKKLRECESVLVN-DFFLVACLEDFIENARLFIFETFCRIHQ 90
P+ E++E Y + +D +L ES D +L + R +E F ++
Sbjct 275 PVREYLESYY-DCHYDRFFIQLAALESERFKFDRYLSPHFNYYSRGMRHRAYEQFLTPYK 333
Query 91 CISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVMGNNAVSP---YQQVIE 147
+ I+M+A ++ +R + LI +L +ID+ G V+ N+ S Y+ VI+
Sbjct 334 TVRIDMMAKDFGVSRAFIDRELHRLIATGQLQCRIDAVNG-VIEVNHRDSKNHLYKAVIK 392
Query 148 KTKSLSFRSQMLA 160
L R Q LA
Sbjct 393 DGDILLNRIQKLA 405
Lambda K H
0.322 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3022542264
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40