bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3690_orf1
Length=401
Score E
Sequences producing significant alignments: (Bits) Value
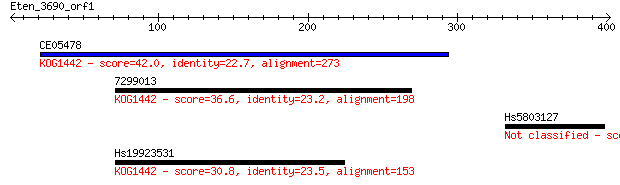
CE05478 42.0 0.002
7299013 36.6 0.089
Hs5803127 31.6 3.6
Hs19923531 30.8 4.9
> CE05478
Length=416
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 62/281 (22%), Positives = 123/281 (43%), Gaps = 24/281 (8%)
Query 21 SLRDVRLPT-WQELQRYVRDPEILTSEILFLILTLSNIFVVYRLFLDI-VPFPVFVTWWQ 78
++R+ PT W+ + +++T+ + + ++ +F+ L + + P+F+TW+Q
Sbjct 67 AMRNRENPTKWESYK------QVITAVSAYWVFSIGLVFLNKYLLSSVQLDAPLFITWYQ 120
Query 79 LAQGLLMAWCLGEVGQEYPKLAYFPRVEIDKRLLRRLFVPTIVNALMLVLANVLLYRVQC 138
+ + L + + Y L FP + ID ++ R + ++V M+ N+ L V
Sbjct 121 CLVTVFLCLFLSKTSKAYG-LFKFPSMPIDAKISREVLPLSVVFVAMISFNNLCLKYVG- 178
Query 139 IATLPVVVAFAVVLHHVTRFVGCGEEYMPMRWQAIG---LLFTAFLLGITDSKTVGSEVL 195
++ V + V + V ++ G++ QAIG L+ FLLG+ G+
Sbjct 179 VSFYYVGRSLTTVFNVVCTYLILGQK---TSGQAIGCCALIIFGFLLGVDQEGVTGTLSY 235
Query 196 PWAL--LYAIFSAAFRAAFLQKVMHEVEGKGNLLHNHQHLISVIILPILMIICGE-GSVL 252
+ + A S A A + +KV+ V L + +L ++++ LM+ GE G+V
Sbjct 236 TGVIFGVLASLSVALNAIYTRKVLSSVGDCLWRLTMYNNLNALVLFLPLMLFNGEFGAVF 295
Query 253 SAMPMNFGSLYTWQTWGCLFTVGALPFLKNIVSNRLIRRTG 293
F L+ W + G F+ V+ I+ T
Sbjct 296 Y-----FDKLFDTTFWILMTLGGVFGFMMGYVTGWQIQATS 331
> 7299013
Length=337
Score = 36.6 bits (83), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 46/210 (21%), Positives = 91/210 (43%), Gaps = 28/210 (13%)
Query 71 PVFVTWWQLAQGLLMAWCLGEVGQEYPKLAYFPRVE-IDKRLLRRLFVPTIVNALMLVLA 129
P+F++W+Q ++ + + ++YP + FP +D R++ +++ LM+
Sbjct 50 PLFMSWFQCVVSTVICFVASRLSRKYPSVFTFPEGNPLDIDTFRKILPLSVLYTLMIGAN 109
Query 130 NVLLYRVQCIATLPVVVAFAVVLHHVTRFVGCGEEYMPMRWQ-------AIGLLFTAFLL 182
N+ L V VAF + +T Y+ +R + G + F L
Sbjct 110 NLSLSY--------VTVAFYYIGRSLTTVFSVVLTYVILRQRTSFKCLLCCGAIVVGFWL 161
Query 183 GITDSKTVGSEVLPW-ALLYAIFSAAFRAAF---LQKVMHEVEGKGNLLHNHQHLISVII 238
G+ D +++ +EV W ++ + S+ A F +K + V + LL + +L S ++
Sbjct 162 GV-DQESL-TEVFSWRGTIFGVLSSLALAMFSIQTKKSLGYVNQEVWLLSYYNNLYSTLL 219
Query 239 LPILMIICGEGSVLSAMPMNFGSLYTWQTW 268
L+II GE + P + W +W
Sbjct 220 FLPLIIINGELESIITYP------HLWASW 243
> Hs5803127
Length=76
Score = 31.6 bits (70), Expect = 3.6, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 32/66 (48%), Gaps = 8/66 (12%)
Query 332 GRALGACDVLLNAAEAEQNEMRPSGATRHTGDTEGGELERGSNSKQYLQSIEEDSGEAYG 391
GR D+L+++A NE+ A TEG E + S+++Q SGEA G
Sbjct 18 GRRNAIHDILVSSASGNSNELALKLAGLDINKTEGEEDAQRSSTEQ--------SGEAQG 69
Query 392 ELAEGE 397
E A+ E
Sbjct 70 EAAKSE 75
> Hs19923531
Length=351
Score = 30.8 bits (68), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 36/164 (21%), Positives = 66/164 (40%), Gaps = 21/164 (12%)
Query 71 PVFVTWWQLAQGLLMAWCLGEVGQEYPKLAYFPRVEIDKRLLRRLFVPTIVNALMLVLAN 130
P+FVT++Q L+ L + P FP + +D R+ R + ++V M+ N
Sbjct 60 PIFVTFYQCLVTTLLCKGLSALAACCPGAVDFPSLRLDLRVARSVLPLSVVFIGMITFNN 119
Query 131 VLLYRVQCIATLPVVVAFAVVLHHVTRFVGCGEEYMPMRWQ-------AIGLLFTAFLLG 183
+ L V VAF V +T Y+ ++ G++ F LG
Sbjct 120 LCLKYVG--------VAFYNVGRSLTTVFNVLLSYLLLKQTTSFYALLTCGIIIGGFWLG 171
Query 184 ITDSKTVGSEVLPW-ALLYAIFSA---AFRAAFLQKVMHEVEGK 223
+ G+ L W ++ + ++ + A + KV+ V+G
Sbjct 172 VDQEGAEGT--LSWLGTVFGVLASLCVSLNAIYTTKVLPAVDGS 213
Lambda K H
0.327 0.141 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10140246140
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40