bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3682_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
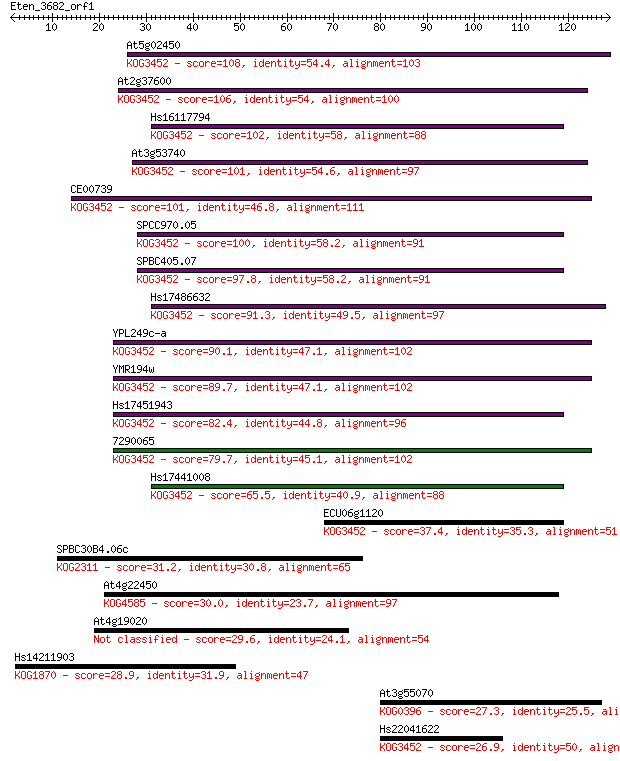
At5g02450 108 2e-24
At2g37600 106 1e-23
Hs16117794 102 2e-22
At3g53740 101 4e-22
CE00739 101 4e-22
SPCC970.05 100 8e-22
SPBC405.07 97.8 4e-21
Hs17486632 91.3 4e-19
YPL249c-a 90.1 9e-19
YMR194w 89.7 1e-18
Hs17451943 82.4 2e-16
7290065 79.7 1e-15
Hs17441008 65.5 2e-11
ECU06g1120 37.4 0.008
SPBC30B4.06c 31.2 0.45
At4g22450 30.0 1.0
At4g19020 29.6 1.4
Hs14211903 28.9 2.3
At3g55070 27.3 8.2
Hs22041622 26.9 8.4
> At5g02450
Length=108
Score = 108 bits (271), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 56/103 (54%), Positives = 75/103 (72%), Gaps = 3/103 (2%)
Query 26 STGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLIKI 85
+TG+ VGLNKG +VTKR PPRP+ RKGK + R IR +I++VAG APYE+R+ +L+K+
Sbjct 3 ATGLFVGLNKGHVVTKREQPPRPNNRKGKTSKRTIFIRNLIKEVAGQAPYEKRITELLKV 62
Query 86 GSAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMKKKAAA 128
G KRALK AK++LGTHKR K KREE+S++ M+ A
Sbjct 63 GK---DKRALKVAKRKLGTHKRAKRKREEMSSVLRKMRSGGAG 102
> At2g37600
Length=113
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 54/100 (54%), Positives = 74/100 (74%), Gaps = 3/100 (3%)
Query 24 STSTGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLI 83
+ TG+ VGLNKG +VT+R PRP+ RKGK + R IR++IR+VAG APYE+R+ +L+
Sbjct 5 AVKTGLFVGLNKGHVVTRRELAPRPNSRKGKTSKRTIFIRKLIREVAGMAPYEKRITELL 64
Query 84 KIGSAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMK 123
K+G KRALK AK++LGTHKR K KREE+S++ M+
Sbjct 65 KVGK---DKRALKVAKRKLGTHKRAKRKREEMSSVLRKMR 101
> Hs16117794
Length=105
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 66/88 (75%), Gaps = 3/88 (3%)
Query 31 VGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLIKIGSAAT 90
VGLNKG VTK + PR SRR+G+ T + +R++IR+V GFAPYERR ++L+K+ +
Sbjct 9 VGLNKGHKVTKNVSKPRHSRRRGRLTKHTKFVRDMIREVCGFAPYERRAMELLKV---SK 65
Query 91 TKRALKFAKKRLGTHKRGKAKREELSNI 118
KRALKF KKR+GTH R K KREELSN+
Sbjct 66 DKRALKFIKKRVGTHIRAKRKREELSNV 93
> At3g53740
Length=112
Score = 101 bits (251), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 53/97 (54%), Positives = 71/97 (73%), Gaps = 3/97 (3%)
Query 27 TGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLIKIG 86
TG+ VGLNKG +VT+R PRP RKGK + R IR +I++VAG APYE+R+ +L+K+G
Sbjct 8 TGLFVGLNKGHVVTRRELAPRPRSRKGKTSKRTIFIRNLIKEVAGQAPYEKRITELLKVG 67
Query 87 SAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMK 123
KRALK AK++LGTHKR K KREE+S++ M+
Sbjct 68 K---DKRALKVAKRKLGTHKRAKRKREEMSSVLRKMR 101
> CE00739
Length=142
Score = 101 bits (251), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 52/113 (46%), Positives = 76/113 (67%), Gaps = 5/113 (4%)
Query 14 FIPIRSLSKMSTS--TGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAG 71
F+PI + + MS G+ VGLNKG T+ R +R KG + + + +RE++R++ G
Sbjct 30 FLPIYAANLMSGPGIEGLAVGLNKGHAATQLPVKQRQNRHKGVASKKTKIVRELVREITG 89
Query 72 FAPYERRVLDLIKIGSAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMKK 124
FAPYERRVL++++I + KRALKF K+R+GTH+R K KREEL N+ A +K
Sbjct 90 FAPYERRVLEMLRI---SKDKRALKFLKRRIGTHRRAKGKREELQNVIIAQRK 139
> SPCC970.05
Length=99
Score = 100 bits (248), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 53/91 (58%), Positives = 68/91 (74%), Gaps = 3/91 (3%)
Query 28 GIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLIKIGS 87
G+ VGLNKG ++TKR P RPSRRKG+ + R +R I+R+VAGFAPYERRV++LI+
Sbjct 4 GLVVGLNKGKVLTKRQLPERPSRRKGQLSKRTSFVRSIVREVAGFAPYERRVMELIR--- 60
Query 88 AATTKRALKFAKKRLGTHKRGKAKREELSNI 118
+ KRA K AKKRLGT KR K K EEL+++
Sbjct 61 NSQDKRARKLAKKRLGTLKRAKGKIEELTSV 91
> SPBC405.07
Length=99
Score = 97.8 bits (242), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 53/91 (58%), Positives = 66/91 (72%), Gaps = 3/91 (3%)
Query 28 GIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLIKIGS 87
G+ VGLNKG +TKR P RPSRRKG + R +R I+R+VAGFAPYERRV++LI+
Sbjct 4 GLVVGLNKGKTLTKRQLPERPSRRKGHLSKRTAFVRSIVREVAGFAPYERRVMELIR--- 60
Query 88 AATTKRALKFAKKRLGTHKRGKAKREELSNI 118
+ KRA K AKKRLGT KR K K EEL+++
Sbjct 61 NSQDKRARKLAKKRLGTLKRAKGKIEELTSV 91
> Hs17486632
Length=105
Score = 91.3 bits (225), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 48/97 (49%), Positives = 65/97 (67%), Gaps = 3/97 (3%)
Query 31 VGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLIKIGSAAT 90
VGLNKG +TK + PR SR G+ T + +R +I++V GF PYER ++L+K+ +
Sbjct 9 VGLNKGHKLTKNLSKPRHSRSLGRPTKHTKCVRGMIQEVCGFTPYERCTMELLKV---SK 65
Query 91 TKRALKFAKKRLGTHKRGKAKREELSNIAAAMKKKAA 127
K+ALKF KKR+GTH K KREELSN+ A +K AA
Sbjct 66 DKQALKFIKKRVGTHIHTKRKREELSNVLAITRKVAA 102
> YPL249c-a
Length=100
Score = 90.1 bits (222), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 48/102 (47%), Positives = 70/102 (68%), Gaps = 3/102 (2%)
Query 23 MSTSTGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDL 82
M+ TGI +GLNKG VT+ P+ S +KG + R + +R ++R++AG +PYERR++DL
Sbjct 1 MAVKTGIAIGLNKGKKVTQMTPAPKISYKKGAASNRTKFVRSLVREIAGLSPYERRLIDL 60
Query 83 IKIGSAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMKK 124
I+ + KRA K AKKRLG+ R KAK EE++NI AA ++
Sbjct 61 IR---NSGEKRARKVAKKRLGSFTRAKAKVEEMNNIIAASRR 99
> YMR194w
Length=100
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/102 (47%), Positives = 69/102 (67%), Gaps = 3/102 (2%)
Query 23 MSTSTGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDL 82
M+ TGI +GLNKG VT P+ S +KG + R + +R ++R++AG +PYERR++DL
Sbjct 1 MTVKTGIAIGLNKGKKVTSMTPAPKISYKKGAASNRTKFVRSLVREIAGLSPYERRLIDL 60
Query 83 IKIGSAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMKK 124
I+ + KRA K AKKRLG+ R KAK EE++NI AA ++
Sbjct 61 IR---NSGEKRARKVAKKRLGSFTRAKAKVEEMNNIIAASRR 99
> Hs17451943
Length=105
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/96 (44%), Positives = 64/96 (66%), Gaps = 3/96 (3%)
Query 23 MSTSTGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDL 82
M+ + +GL KG VTK + PR S R+ + T + + ++IR+V GFAPY+RR ++L
Sbjct 1 MALGYPMAMGLFKGHKVTKNVSKPRHSSRRRRLTKHTKFVWDMIREVCGFAPYKRRAMEL 60
Query 83 IKIGSAATTKRALKFAKKRLGTHKRGKAKREELSNI 118
+K+ + K+ALKF KKR+G H R K K+EELSN+
Sbjct 61 LKV---SKDKQALKFIKKRVGMHIRAKRKQEELSNV 93
> 7290065
Length=115
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 46/112 (41%), Positives = 67/112 (59%), Gaps = 13/112 (11%)
Query 23 MSTSTGIRVGLNKG----------FLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGF 72
M+ + +GLNKG + K+ R SR K +T + +R+++R+V G
Sbjct 1 MAVRYELAIGLNKGHKTSKIRNVKYTGDKKVKGLRGSRLKNIQTRHTKFMRDLVREVVGH 60
Query 73 APYERRVLDLIKIGSAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMKK 124
APYE+R ++L+K+ + KRALKF K+RLGTH R K KREELSNI ++K
Sbjct 61 APYEKRTMELLKV---SKDKRALKFLKRRLGTHIRAKRKREELSNILTQLRK 109
> Hs17441008
Length=100
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 50/88 (56%), Gaps = 8/88 (9%)
Query 31 VGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVLDLIKIGSAAT 90
+GLNKG VTK + PR S + T + ++++IR+V GFAPYE ++L K+ A
Sbjct 9 MGLNKGHKVTKNVSNPRHSCHRRCLTKHTKFVQDMIREVCGFAPYECHTMELQKVSKAME 68
Query 91 TKRALKFAKKRLGTHKRGKAKREELSNI 118
+ KF KKR KREELSN+
Sbjct 69 LLKTSKFIKKR--------RKREELSNV 88
> ECU06g1120
Length=107
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 32/51 (62%), Gaps = 3/51 (5%)
Query 68 QVAGFAPYERRVLDLIKIGSAATTKRALKFAKKRLGTHKRGKAKREELSNI 118
+++G +P E++ + L+ A +A K +KRLG+HKR AK E+L+ +
Sbjct 56 EISGLSPLEKKAISLL---EAKNNNKAQKLLRKRLGSHKRAVAKVEKLARM 103
> SPBC30B4.06c
Length=666
Score = 31.2 bits (69), Expect = 0.45, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 32/65 (49%), Gaps = 1/65 (1%)
Query 11 PLVFIPIRSLSKMSTSTGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVA 70
P + PI ++SKM+ TG + FL +R P +R +T + EI+R+
Sbjct 219 PRLSSPI-NISKMTEQTGDEIPETFSFLNLERDFSPALPQRSCYRTYTTELTHEIVRKNL 277
Query 71 GFAPY 75
FAP+
Sbjct 278 AFAPH 282
> At4g22450
Length=457
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 23/97 (23%), Positives = 42/97 (43%), Gaps = 4/97 (4%)
Query 21 SKMSTSTGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGFAPYERRVL 80
SK STS + + N ++ R PP SR + R ++R +I + G + R+L
Sbjct 315 SKQSTSQSVVLQENVSLMLFNRGGPPTNSRELFNR--RHSSLRSVIERTFGVWKAKWRIL 372
Query 81 DLIKIGSAATTKRALKFAKKRLGTHKRGKAKREELSN 117
D + K+ +K + H + ++E S+
Sbjct 373 D--RKHPKYEVKKWIKIVTSTMALHNYIRDSQQEDSD 407
> At4g19020
Length=1171
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 13/54 (24%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 19 SLSKMSTSTGIRVGLNKGFLVTKRATPPRPSRRKGKKTTRVQAIREIIRQVAGF 72
+L+++ + + + G N+G + P + ++RKGK+ TR + R + F
Sbjct 470 NLTQVESGSALSSGGNEGIVSLDLNNPTKSTKRKGKRVTRTAVQEQNKRSICFF 523
> Hs14211903
Length=1274
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 20/47 (42%), Gaps = 0/47 (0%)
Query 2 TALLCNFIIPLVFIPIRSLSKMSTSTGIRVGLNKGFLVTKRATPPRP 48
+ LC F IP+ PI + S T N+ F +T PRP
Sbjct 659 SGFLCAFEIPVPVSPISASSPTQTDFSSSPSTNEMFTLTTNGDLPRP 705
> At3g55070
Length=418
Score = 27.3 bits (59), Expect = 8.2, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 30/47 (63%), Gaps = 2/47 (4%)
Query 80 LDLIKIGSAATTKRALKFAKKRLGTHKRGKAKREELSNIAAAMKKKA 126
++L+++ +A + K+A+++A+K L + G +EL ++ A + K+
Sbjct 226 IELVRVDTAESYKKAIQYARKHLASW--GTTHMKELQHVLATLAFKS 270
> Hs22041622
Length=108
Score = 26.9 bits (58), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 3/26 (11%)
Query 80 LDLIKIGSAATTKRALKFAKKRLGTH 105
++L+K+ + K ALKF KKR+GTH
Sbjct 1 MELLKV---SKDKWALKFIKKRVGTH 23
Lambda K H
0.322 0.134 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40