bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
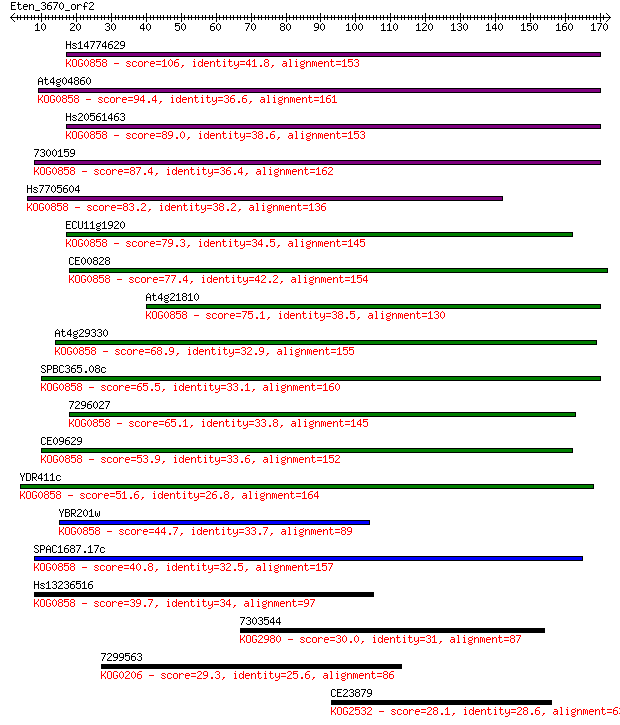
Query= Eten_3670_orf2
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
Hs14774629 106 2e-23
At4g04860 94.4 1e-19
Hs20561463 89.0 4e-18
7300159 87.4 1e-17
Hs7705604 83.2 3e-16
ECU11g1920 79.3 3e-15
CE00828 77.4 1e-14
At4g21810 75.1 6e-14
At4g29330 68.9 5e-12
SPBC365.08c 65.5 4e-11
7296027 65.1 7e-11
CE09629 53.9 1e-07
YDR411c 51.6 8e-07
YBR201w 44.7 9e-05
SPAC1687.17c 40.8 0.001
Hs13236516 39.7 0.003
7303544 30.0 2.1
7299563 29.3 4.3
CE23879 28.1 9.1
> Hs14774629
Length=239
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 64/157 (40%), Positives = 89/157 (56%), Gaps = 6/157 (3%)
Query 17 HIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFSLHF 76
IPPV+R A + A LE+I+PF LY N +L+F H Q+WRLIT FLFFG +F
Sbjct 12 QIPPVSRAYTTACVLTTAAVQLELITPFQLYFNPELIFKHFQIWRLITNFLFFGPVGFNF 71
Query 77 FWNVYVLIFYCSTLEE---HRRSATFLWMLICTGGLLLAFSYIFGVSSYFFSGSMINVMT 133
+N+ L YC LEE R+A F++M + G L+ F VS F + ++
Sbjct 72 LFNMIFLYRYCRMLEEGSFRGRTADFVFMFLFGGFLMTLFGLF--VSLVFLGQAFTIMLV 129
Query 134 YIWGRRNPNTRLSIF-FMPVLAPYLPFLLALVSLLGG 169
Y+W RRNP R++ F + AP+LP++L SLL G
Sbjct 130 YVWSRRNPYVRMNFFGLLNFQAPFLPWVLMGFSLLLG 166
> At4g04860
Length=244
Score = 94.4 bits (233), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 59/174 (33%), Positives = 93/174 (53%), Gaps = 13/174 (7%)
Query 9 EQVDLFLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLF 68
+ V+ + +P +TR L A+ + C+L+IISP++LY+N LV Q WRL+T FL+
Sbjct 3 QAVEEWYKQMPIITRSYLTAAVITTVGCSLDIISPYNLYLNPTLVVKQYQYWRLVTNFLY 62
Query 69 FGTFSLHFFWNVYVLIFYCSTLEEHR---RSATFLWMLICTGGLLLAFSYIFGVSSY--- 122
F L F ++++ L YC LEE+ ++A FL+ML+ +L I G+ Y
Sbjct 63 FRKMDLDFMFHMFFLARYCKLLEENSFRGKTADFLYMLLFGASVLTGIVLIGGMIPYLSA 122
Query 123 ------FFSGSMINVMTYIWGRRNPNTRLSIF-FMPVLAPYLPFLLALVSLLGG 169
F S S+ +M Y+W ++NP +S A YLP++L S+L G
Sbjct 123 SFAKIIFLSNSLTFMMVYVWSKQNPYIHMSFLGLFTFTAAYLPWVLLGFSILVG 176
> Hs20561463
Length=239
Score = 89.0 bits (219), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 59/157 (37%), Positives = 85/157 (54%), Gaps = 6/157 (3%)
Query 17 HIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFSLHF 76
+P VTR A + A LE++SPF LY N LVF QVWRL+T FLFFG F
Sbjct 12 QVPAVTRAYTAACVLTTAAVQLELLSPFQLYFNPHLVFRKFQVWRLVTNFLFFGPLGFSF 71
Query 77 FWNVYVLIFYCSTLEE---HRRSATFLWMLICTGGLLLAFSYIFGVSSYFFSGSMINVMT 133
F+N+ + YC LEE R+A F++M + +L S +F +++ ++
Sbjct 72 FFNMLFVFRYCRMLEEGSFRGRTADFVFMFLFG--GVLMTLLGLLGSLFFLGQALMAMLV 129
Query 134 YIWGRRNPNTRLSIF-FMPVLAPYLPFLLALVSLLGG 169
Y+W RR+P R++ F + AP+LP+ L SLL G
Sbjct 130 YVWSRRSPRVRVNFFGLLTFQAPFLPWALMGFSLLLG 166
> 7300159
Length=261
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 59/166 (35%), Positives = 87/166 (52%), Gaps = 6/166 (3%)
Query 8 MEQVDLFLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFL 67
M + F IP VTR + L+++SP LY N L+ Q+WRL T FL
Sbjct 1 MNALRQFYLEIPVVTRAYTTVCVLTTLAVHLDLVSPLQLYFNPTLIVRKFQIWRLATTFL 60
Query 68 FFGTFSLHFFWNVYVLIFYCSTLEE---HRRSATFLWMLICTGGLLLAFSYIFGVSSYFF 124
+FGT + FF+N+ YC LE+ RS+ F+ M I GG+L+ F IF V+ F
Sbjct 61 YFGTIGISFFFNMVFTYRYCRMLEDGSFRGRSSDFVMMFIF-GGVLMTFFGIF-VNLLFL 118
Query 125 SGSMINVMTYIWGRRNPNTRLSIF-FMPVLAPYLPFLLALVSLLGG 169
+ ++ Y+W RRNP ++ F + APYLP++L S++ G
Sbjct 119 GQAFTLMLVYVWSRRNPLVPMNFFGVLNFQAPYLPWVLLCCSMILG 164
> Hs7705604
Length=209
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 52/139 (37%), Positives = 74/139 (53%), Gaps = 5/139 (3%)
Query 6 LKMEQVDLFLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITC 65
+ + + L IPPV+R A + A LE+I+PF LY N +L+F H Q+WRLIT
Sbjct 1 MAYQSLRLEYLQIPPVSRAYTTACVLTTAAVQLELITPFQLYFNPELIFKHFQIWRLITN 60
Query 66 FLFFGTFSLHFFWNVYVLIFYCSTLEE---HRRSATFLWMLICTGGLLLAFSYIFGVSSY 122
FLFFG +F +N+ L YC LEE R+A F++M + GG L+ +F VS
Sbjct 61 FLFFGPVGFNFLFNMIFLYRYCRMLEEGSFRGRTADFVFMFL-FGGFLMTLFGLF-VSLV 118
Query 123 FFSGSMINVMTYIWGRRNP 141
F + N + + G P
Sbjct 119 FLGPGLYNNGSSMCGAEEP 137
> ECU11g1920
Length=348
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 50/147 (34%), Positives = 81/147 (55%), Gaps = 5/147 (3%)
Query 17 HIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFSLHF 76
+PP+TR + + + L ++ +SP+SLY + L ++WR+ T FL+FG +L
Sbjct 18 RVPPITRYMTLLISAVALLVYVDAVSPYSLYYS-PLFLKRLEIWRVFTSFLYFGKPTLDM 76
Query 77 FWNVYVLIFYCSTLEEH-RRSATFLWMLICTGGLLLAFSYIFGVSSYFFSGSMINVMTYI 135
F +V L Y LEE ++ + W+++ +L A S I+G+S+ S +TYI
Sbjct 77 FMHVVFLYRYSRMLEEGCVNTSEYFWLILVISSVLFAISNIYGISA--LGTSFSATITYI 134
Query 136 WGRRNPNTRLSIF-FMPVLAPYLPFLL 161
W +RNP + IF F+ A YLPF+L
Sbjct 135 WTKRNPRAIVQIFGFISFPAFYLPFIL 161
> CE00828
Length=227
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 65/158 (41%), Positives = 88/158 (55%), Gaps = 6/158 (3%)
Query 18 IPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFSLHFF 77
+PPVTR A +L LE ++PF LY NW+L+ Q WRLIT F FFG+F F
Sbjct 1 MPPVTRFYTGACVLLTTAVHLEFVTPFHLYFNWELIIRKYQFWRLITSFCFFGSFGFSFL 60
Query 78 WNVYVLIFYCSTLEE---HRRSATFLWMLICTGGLLLAFSYIFGVSSYFFSGSMINVMTY 134
+N+ YC LEE R A F++M + G +L+ S IF V F + ++ Y
Sbjct 61 FNMIFTYRYCMMLEEGSFRGRRADFVYMFLF-GAVLMILSGIF-VQILFLGQAFTIMLVY 118
Query 135 IWGRRNPNTRLSIF-FMPVLAPYLPFLLALVSLLGGWN 171
IW RRNP +++ F + APYLP++L L SLL G N
Sbjct 119 IWSRRNPMIQMNFFGVLTFTAPYLPWVLLLFSLLLGNN 156
> At4g21810
Length=225
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 50/143 (34%), Positives = 75/143 (52%), Gaps = 13/143 (9%)
Query 40 IISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFSLHFFWNVYVLIFYCSTLEEHR---RS 96
IISP++LY+N LV Q WRL+T FL+F L F ++++ L YC LEE+ ++
Sbjct 15 IISPYNLYLNPTLVVKQYQFWRLVTNFLYFRKMDLDFLFHMFFLARYCKLLEENSFRGKT 74
Query 97 ATFLWMLICTGGLLLAFSYIFGVSSY---------FFSGSMINVMTYIWGRRNPNTRLSI 147
FL+ML+ +L I G+ Y F S S+ +M Y+W ++NP +S
Sbjct 75 TDFLYMLLFGATVLTGIVLIGGMIPYLSVSFSKIIFLSNSLTFMMVYVWSKQNPYIHMSF 134
Query 148 F-FMPVLAPYLPFLLALVSLLGG 169
A YLP++L S+L G
Sbjct 135 LGLFTFTAAYLPWVLLGFSILVG 157
> At4g29330
Length=281
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 82/160 (51%), Gaps = 5/160 (3%)
Query 14 FLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFS 73
F +PP+T+ L +++P + + +LV Q+WRLIT F G FS
Sbjct 22 FYNSLPPITKAYGTLCFFTTVATQLGLVAPVHIALIPELVLKQFQIWRLITNLFFLGGFS 81
Query 74 LHFFWNVYVLIFYCSTLEE---HRRSATFLWMLICTGGLLLAFSYIFGVSSYFFSGSMIN 130
++F + ++ Y LE+ RR+A FLWM+I LL S I + F S++
Sbjct 82 INFGIRLLMIARYGVQLEKGPFERRTADFLWMMIFGSFTLLVLSVIPFFWTPFLGVSLVF 141
Query 131 VMTYIWGRRNPNTRLSIFFMPVL-APYLPF-LLALVSLLG 168
++ Y+W R PN +S++ + L A YLP+ +LAL + G
Sbjct 142 MLLYLWSREFPNANISLYGLVTLKAFYLPWAMLALDVIFG 181
> SPBC365.08c
Length=224
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 53/166 (31%), Positives = 84/166 (50%), Gaps = 10/166 (6%)
Query 10 QVDLFLTHIPPVTRVCLV--ASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFL 67
Q+ L+ IPPVTR L+ A+T ++ LC +++SP L +++ LV Q +RL T +L
Sbjct 8 QIQELLSRIPPVTRYILLGTAATTILTLC--QLLSPSMLVLHYPLVVRQKQWYRLFTNYL 65
Query 68 FFGTFSLHFFWNVYVLIFYCSTLEE---HRRSATFLWMLICTGGLLLAFSYIFGVSSYFF 124
+ GT F N+Y Y + LE R + ++ L+ L+ AFS I G+ S
Sbjct 66 YAGT-GFDFIMNIYFFYQYSTYLENFVFARNAKKYIIYLVKVALLIDAFSLISGLGSA-L 123
Query 125 SGSMINVMTYIWGRRNPNTRLSIFF-MPVLAPYLPFLLALVSLLGG 169
+ S+ + Y W N +++ F V YLP++L S L G
Sbjct 124 NQSLAAAIAYNWSLFNSFSKIQFLFGFHVQGKYLPYVLLGFSFLTG 169
> 7296027
Length=245
Score = 65.1 bits (157), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 49/152 (32%), Positives = 78/152 (51%), Gaps = 9/152 (5%)
Query 18 IPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFF---GTFSL 74
+P TR L A+ VL LC ++I L+++ VF+ Q+WR +T F +
Sbjct 10 LPRFTRYWLTATVVLSMLCRFDVIPLHWLHLDRSAVFSKLQLWRCMTSLFVFPISSNTAF 69
Query 75 HFFWNVYVLIFYCSTLEEH---RRSATFLWMLICTGGLLLAFSYIFGVSSYFFSGSMINV 131
HF N + ++ Y S LE+ R A +L++LI + L IF V YF +++
Sbjct 70 HFLINCFFIVQYSSKLEKDQYSRSPADYLYLLIVSAVLANIGGMIFNV--YFLMDTLVLA 127
Query 132 MTYIWGRRNPNTRLSIFF-MPVLAPYLPFLLA 162
+TYIW + N + +S +F A YLP++LA
Sbjct 128 ITYIWCQLNKDVTVSFWFGTRFKAMYLPWVLA 159
> CE09629
Length=245
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 51/164 (31%), Positives = 79/164 (48%), Gaps = 19/164 (11%)
Query 10 QVDLFLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFF 69
++ FL IP VTR +AST++ L I+ +++ W LV Q WR +T +++
Sbjct 2 DLENFLLGIPIVTRYWFLASTIIPLLGRFGFINVQWMFLQWDLVVNKFQFWRPLTALIYY 61
Query 70 ---GTFSLHFFWNVYVLIFYCSTLEEHR---RSATFLWMLI-----CTGGLLLAFSYIFG 118
H+ Y L Y LE RSA +L+MLI C+ GL +A
Sbjct 62 PVTPQTGFHWLMMCYFLYNYSKALESETYRGRSADYLFMLIFNWFFCS-GLCMA------ 114
Query 119 VSSYFFSGSMINVMTYIWGRRNPNTRLSIFF-MPVLAPYLPFLL 161
+ YF M+ + Y+W + N +T +S +F M A YLP++L
Sbjct 115 LDIYFLLEPMVISVLYVWCQVNKDTIVSFWFGMRFPARYLPWVL 158
> YDR411c
Length=341
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 44/186 (23%), Positives = 78/186 (41%), Gaps = 24/186 (12%)
Query 4 HGLKMEQVDL-----FLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQ 58
HG D+ F +IPP+TR + V+ + L +I+P+ W L F Q
Sbjct 11 HGNGGRNNDVMGPKEFWLNIPPITRTLFTLAIVMTIVGRLNLINPWYFIYVWNLTFKKVQ 70
Query 59 VWRLITCFLFFGTFSLHFFWNVYVLIFYCSTLEE--------HRRSATFL-----WMLIC 105
+WRL+T + + ++ +Y + S LE +RR ++ C
Sbjct 71 IWRLLTSCVMLSSRAMPALMELYSIYDRSSQLERGHFGPGLSNRRGPMVTVDYAYYLCFC 130
Query 106 TGGLLLAFSYIFGVSSYF---FSGSMINVMTYIWGRRNPNTRLSIF-FMPVLAPYLPFLL 161
+ A + I+G SY+ + I+ +TY W N N ++ + +PV Y P +
Sbjct 131 ILAITTATTIIYG--SYYPVVLTSGFISCITYTWSIDNANVQIMFYGLIPVWGKYFPLIQ 188
Query 162 ALVSLL 167
+S +
Sbjct 189 LFISFV 194
> YBR201w
Length=211
Score = 44.7 bits (104), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Query 15 LTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFSL 74
L IP VTR+ + VL L +L I+ P + ++ LVF Q RL+ +G F+
Sbjct 9 LGDIPLVTRLWTIGCLVLSGLTSLRIVDPGKVVYSYDLVFKKGQYGRLLYSIFDYGAFNW 68
Query 75 HFFWNVYVLIFYCSTLEEH---RRSATFLWML 103
N++V + STLE RR F W++
Sbjct 69 ISMINIFVSANHLSTLENSFNLRRK--FCWII 98
> SPAC1687.17c
Length=168
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 51/164 (31%), Positives = 77/164 (46%), Gaps = 34/164 (20%)
Query 8 MEQVDLFLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFL 67
M + F++ PPVTR +V T+ L A WR IT FL
Sbjct 1 MAILPEFISQTPPVTR-YIVLGTLFTTL---------------------AVYWRAITTFL 38
Query 68 FFGTFSLHFFWNVYVLIFYCSTLEEHR---RSATFLW-MLICTGGLLLA--FSYIFGVSS 121
+ G F L + L+ + S LE ++ +FL +LI LL+ FSY+ +S
Sbjct 39 YVGPFGLELILYLSFLLRFMSMLERSSPPPQTQSFLKTVLIVWFSLLVTSYFSYMPFAAS 98
Query 122 YFFSGSMINVMTYIWGRRNPNTRLSIF-FMPVLAPYLPFLLALV 164
Y FS +M+ YIW ++P R+SI V APY+P+++ L+
Sbjct 99 Y-FSFTML----YIWSWKHPLYRISILGLFDVKAPYVPWVMVLL 137
> Hs13236516
Length=251
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 51/105 (48%), Gaps = 10/105 (9%)
Query 8 MEQVDLFLTHIPPVTRVCLVASTVLMALCTLEIISPFSLYMNWQLVFAHA-QVWRLITCF 66
M + + IP +TR A+ + + L +ISP L++ W F + Q+WR IT
Sbjct 1 MSDIGDWFRSIPAITRYWFAATVAVPLVGKLGLISPAYLFL-WPEAFLYRFQIWRPITAT 59
Query 67 LFF----GTFSLHFFWNVYVLIFYCSTLEE---HRRSATFLWMLI 104
+F GT L+ N+Y L Y + LE R A +L+ML+
Sbjct 60 FYFPVGPGTGFLYLV-NLYFLYQYSTRLETGAFDGRPADYLFMLL 103
> 7303544
Length=351
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 47/97 (48%), Gaps = 11/97 (11%)
Query 67 LFFGTFS----LHFFWNVYVLIFYCSTLEEHRRSATFLWMLICTG--GLLLAFSYIFGVS 120
+F TFS +H F N+YV+ + + FL + + G L++ Y S
Sbjct 188 MFLSTFSHYSAMHLFANMYVMHSFANAAAVSLGKEQFLAVYLSAGVFSSLMSVLYKAATS 247
Query 121 ----SYFFSGSMINVMTYIWGRRNPNTRLSIFFMPVL 153
S SG+++ ++ Y+ + P+T+LSI F+P L
Sbjct 248 QAGMSLGASGAIMTLLAYVC-TQYPDTQLSILFLPAL 283
> 7299563
Length=1718
Score = 29.3 bits (64), Expect = 4.3, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 40/86 (46%), Gaps = 9/86 (10%)
Query 27 VASTVLMALCTLEIISPFSLYMNWQLVFAHAQVWRLITCFLFFGTFSLHFFWNVYVLIFY 86
V +T+L+ T +I SLY ++ V H +W + + F L +F+N + Y
Sbjct 1472 VVATILIVDNTAQI----SLYTSYWTVVNHVTIWGSLVWY-----FVLDYFYNYVIGGPY 1522
Query 87 CSTLEEHRRSATFLWMLICTGGLLLA 112
+L + + TF ++ T L+A
Sbjct 1523 VGSLTQAMKDLTFWVTMLITVMALVA 1548
> CE23879
Length=487
Score = 28.1 bits (61), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 32/68 (47%), Gaps = 18/68 (26%)
Query 93 HRRSATFLWMLICTGGLLLAFSYIFGVSSY-----FFSGSMINVMTYIWGRRNPNTRLSI 147
+R+ T+L+ + G + F I G+S+Y FF+ M+ T L+
Sbjct 85 NRQEKTWLFSTVAVGAMFGLFPVIIGISTYGLRKVFFAAGML-------------TSLTT 131
Query 148 FFMPVLAP 155
F +P++AP
Sbjct 132 FLIPIVAP 139
Lambda K H
0.335 0.144 0.489
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40