bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3647_orf2
Length=197
Score E
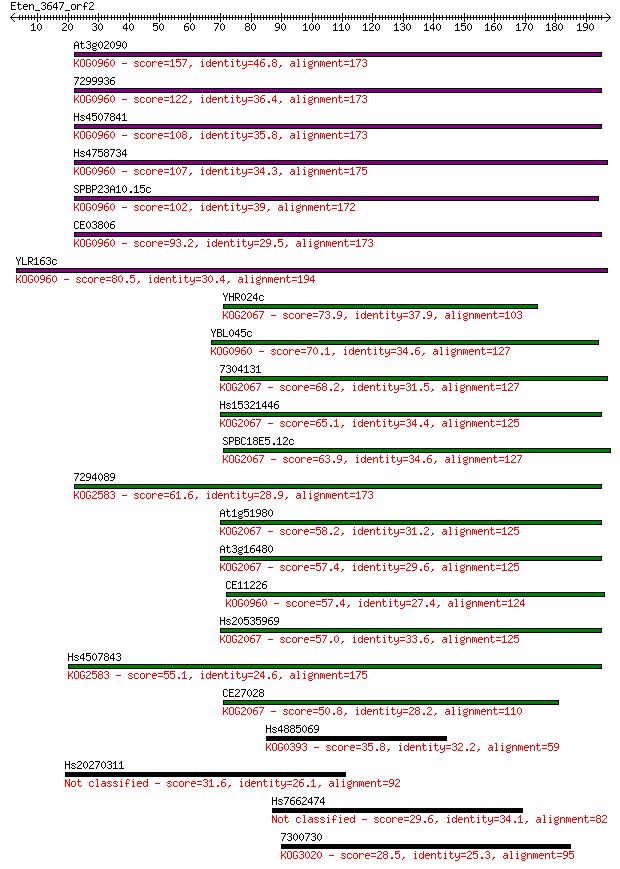
Sequences producing significant alignments: (Bits) Value
At3g02090 157 1e-38
7299936 122 4e-28
Hs4507841 108 8e-24
Hs4758734 107 1e-23
SPBP23A10.15c 102 5e-22
CE03806 93.2 3e-19
YLR163c 80.5 2e-15
YHR024c 73.9 2e-13
YBL045c 70.1 2e-12
7304131 68.2 1e-11
Hs15321446 65.1 8e-11
SPBC18E5.12c 63.9 2e-10
7294089 61.6 1e-09
At1g51980 58.2 1e-08
At3g16480 57.4 2e-08
CE11226 57.4 2e-08
Hs20535969 57.0 2e-08
Hs4507843 55.1 9e-08
CE27028 50.8 2e-06
Hs4885069 35.8 0.058
Hs20270311 31.6 1.2
Hs7662474 29.6 3.6
7300730 28.5 8.0
> At3g02090
Length=531
Score = 157 bits (397), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 81/177 (45%), Positives = 116/177 (65%), Gaps = 14/177 (7%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D VA M+MQ ++GS+ K+ G GK + + +A AE AFNT Y+DTGL
Sbjct 351 DSVALMVMQTMLGSWNKNVGG---GKHVGSDLTQRVAI---NEIAESIMAFNTNYKDTGL 404
Query 82 FGFYAQCDEVAVEHCVLDL----MHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSV 137
FG YA VA C+ DL M+ +T L+Y V+D +V RA+ QL + LL H+D T+ +
Sbjct 405 FGVYA----VAKADCLDDLSYAIMYEVTKLAYRVSDADVTRARNQLKSSLLLHMDGTSPI 460
Query 138 AEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
AED+GRQ+L YGR +PTAE R++A+DA +KRVA KY++DK++A+SA+GP+ +P
Sbjct 461 AEDIGRQLLTYGRRIPTAELFARIDAVDASTVKRVANKYIYDKDIAISAIGPIQDLP 517
> 7299936
Length=382
Score = 122 bits (307), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 63/173 (36%), Positives = 98/173 (56%), Gaps = 6/173 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D + M+ ++G++ + + G G +A+ R A + F +FNTCY+DTGL
Sbjct 202 DNIPLMVANTLVGAWDRSQGG---GANNASNLARASA---EDNLCHSFQSFNTCYKDTGL 255
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
+G Y CD + E + ++ L VT+ EVERAK L T +L LD TT + ED+
Sbjct 256 WGIYFVCDPLQCEDMLFNVQTEWMRLCTMVTEAEVERAKNLLKTNMLLQLDGTTPICEDI 315
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
GRQIL Y R +P E R++A+ ++ VA KY++D+ AV+A+GP+ +P
Sbjct 316 GRQILCYNRRIPLHELEQRIDAVSVGNVRDVAMKYIYDRCPAVAAVGPVENLP 368
> Hs4507841
Length=480
Score = 108 bits (269), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 62/173 (35%), Positives = 91/173 (52%), Gaps = 6/173 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D VA + AIIG Y G V LS+ +A K+ + F F+ CY +TGL
Sbjct 300 DSVALQVANAIIGHYDCTYGGGV--HLSSPLASGAVANKL----CQSFQTFSICYAETGL 353
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
G + CD + ++ + L L S T+ EV R K L L+ HLD TT V ED+
Sbjct 354 LGAHFVCDRMKIDDMMFVLQGQWMRLCTSATESEVARGKNILRNALVSHLDGTTPVCEDI 413
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
GR +L YGR +P AE+ R+ +DA ++ + KY++D+ AV+ GP+ +P
Sbjct 414 GRSLLTYGRRIPLAEWESRIAEVDASVVREICSKYIYDQCPAVAGYGPIEQLP 466
> Hs4758734
Length=489
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 60/184 (32%), Positives = 100/184 (54%), Gaps = 24/184 (13%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKM-QTGC----AEMFSAFNTCY 76
D + M+ +IG++ + G + N+++K+ Q C F +FNT Y
Sbjct 309 DTICLMVANTLIGNWDRSFGGGM-----------NLSSKLAQLTCHGNLCHSFQSFNTSY 357
Query 77 RDTGLFGFYAQCDEVAVEHCVLDLMHGITA----LSYSVTDEEVERAKAQLTTQLLGHLD 132
DTGL+G Y C+ V D++H + L SVT+ EV RA+ L T +L LD
Sbjct 358 TDTGLWGLYMVCESSTVA----DMLHVVQKEWMRLCTSVTESEVARARNLLKTNMLLQLD 413
Query 133 STTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHG 192
+T + ED+GRQ+L Y R +P E R++A++A+ I+ V KY++++ A++A+GP+
Sbjct 414 GSTPICEDIGRQMLCYNRRIPIPELEARIDAVNAETIREVCTKYIYNRSPAIAAVGPIKQ 473
Query 193 MPQL 196
+P
Sbjct 474 LPDF 477
> SPBP23A10.15c
Length=457
Score = 102 bits (254), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 67/176 (38%), Positives = 100/176 (56%), Gaps = 14/176 (7%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D ++MQAIIG++ + G P S +T+ Q A F +F+T Y DTGL
Sbjct 277 DYFTALVMQAIIGNWDR-AMGASPHLSSRLSTI-----VQQHQLANSFMSFSTSYSDTGL 330
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYS----VTDEEVERAKAQLTTQLLGHLDSTTSV 137
+G Y + + + DL+H T +++ T EVERAKAQL LL LDSTT++
Sbjct 331 WGIYLVTENLG---RIDDLVH-FTLQNWARLTVATRAEVERAKAQLRASLLLSLDSTTAI 386
Query 138 AEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGM 193
AED+GRQ+L GR + E +R+ I ++ RVA + + DK++AVSA+G + G+
Sbjct 387 AEDIGRQLLTTGRRMSPQEVDLRIGQITEKDVARVASEMIWDKDIAVSAVGSIEGL 442
> CE03806
Length=485
Score = 93.2 bits (230), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 51/173 (29%), Positives = 87/173 (50%), Gaps = 6/173 (3%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
D +A M+ ++G Y + N R Q E+F +FNTCY++TGL
Sbjct 306 DNLALMVANTLMGEYDRMR------GFGVNAPTRLAEKLSQDAGIEVFQSFNTCYKETGL 359
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
G Y ++++ + ++ L+ ++ + V+RAK L T LL LD +T V ED+
Sbjct 360 VGTYFVAAPESIDNLIDSVLQQWVWLANNIDEAAVDRAKRSLHTNLLLMLDGSTPVCEDI 419
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
GRQ+L YGR +PT E R+ +I +++ V + + +V+ + +G P
Sbjct 420 GRQLLCYGRRIPTPELHARIESITVQQLRDVCRRVFLEGQVSAAVVGKTQYWP 472
> YLR163c
Length=462
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 59/205 (28%), Positives = 101/205 (49%), Gaps = 19/205 (9%)
Query 3 FIPQFTSLPVSPLLLLL------CADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRN 56
FI + T LP + + + L D + QAI+G++ + G + + +
Sbjct 257 FIKENT-LPTTHIAIALEGVSWSAPDYFVALATQAIVGNWDR-----AIGTGTNSPSPLA 310
Query 57 IATKMQTGCAEMFSAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITA-----LSYSV 111
+A A + +F+T Y D+GL+G Y D + EH V +++ I S +
Sbjct 311 VAASQNGSLANSYMSFSTSYADSGLWGMYIVTD--SNEHNVQLIVNEILKEWKRIKSGKI 368
Query 112 TDEEVERAKAQLTTQLLGHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKR 171
+D EV RAKAQL LL LD +T++ ED+GRQ++ G+ + E +++ I D+I
Sbjct 369 SDAEVNRAKAQLKAALLLSLDGSTAIVEDIGRQVVTTGKRLSPEEVFEQVDKITKDDIIM 428
Query 172 VAWKYLHDKEVAVSAMGPLHGMPQL 196
A L +K V++ A+G +P +
Sbjct 429 WANYRLQNKPVSMVALGNTSTVPNV 453
> YHR024c
Length=482
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 59/107 (55%), Gaps = 4/107 (3%)
Query 71 AFNTCYRDTGLFGFYAQC----DEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQ 126
AFN Y D+G+FG C AVE + + +T++EV RAK QL +
Sbjct 318 AFNHSYSDSGIFGISLSCIPQAAPQAVEVIAQQMYNTFANKDLRLTEDEVSRAKNQLKSS 377
Query 127 LLGHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVA 173
LL +L+S ED+GRQ+L++GR +P E + ++ + D+I RVA
Sbjct 378 LLMNLESKLVELEDMGRQVLMHGRKIPVNEMISKIEDLKPDDISRVA 424
> YBL045c
Length=457
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 44/134 (32%), Positives = 73/134 (54%), Gaps = 11/134 (8%)
Query 67 EMFSAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMH----GITALSYSVTDEEVERAKAQ 122
+ F+ F+ Y+D+GL+GF V + + DL+H L+ SVTD EVERAK+
Sbjct 313 DNFNHFSLSYKDSGLWGFSTATRNVTM---IDDLIHFTLKQWNRLTISVTDTEVERAKSL 369
Query 123 LTTQLLGHLDSTTSVAED---LGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHD 179
L Q LG L + + D LG ++L+ G + E +++AI ++K A K L D
Sbjct 370 LKLQ-LGQLYESGNPVNDANLLGAEVLIKGSKLSLGEAFKKIDAITVKDVKAWAGKRLWD 428
Query 180 KEVAVSAMGPLHGM 193
+++A++ G + G+
Sbjct 429 QDIAIAGTGQIEGL 442
> 7304131
Length=556
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 40/127 (31%), Positives = 68/127 (53%), Gaps = 1/127 (0%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLG 129
+A+N Y D GLF + + V L + ++ EE+ R+K QL + LL
Sbjct 408 TAYNHAYGDCGLFCVHGSAPPQHMNDMVEVLTREMMGMAAEPGREELMRSKIQLQSMLLM 467
Query 130 HLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGP 189
+L+S V ED+GRQ+LV G+ F+ + ++ A +I+RVA + L +V+A G
Sbjct 468 NLESRPVVFEDVGRQVLVTGQRKRPQHFIKEIESVTAADIQRVAQRLLSSPP-SVAARGD 526
Query 190 LHGMPQL 196
+H +P++
Sbjct 527 IHNLPEM 533
> Hs15321446
Length=525
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 68/126 (53%), Gaps = 3/126 (2%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLG 129
++++ Y DTGL +A D V V + + +V E+ERAK QLT+ L+
Sbjct 378 TSYHHSYEDTGLLCIHASADPRQVREMVEIITKEFILMGGTVDTVELERAKTQLTSMLMM 437
Query 130 HLDSTTSVAEDLGRQIL-VYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L+S + ED+GRQ+L R +P E + + +++KRVA K L K AV+A+G
Sbjct 438 NLESRPVIFEDVGRQVLATRSRKLP-HELCTLIRNVKPEDVKRVASKMLRGKP-AVAALG 495
Query 189 PLHGMP 194
L +P
Sbjct 496 DLTDLP 501
> SPBC18E5.12c
Length=494
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 44/128 (34%), Positives = 65/128 (50%), Gaps = 4/128 (3%)
Query 71 AFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGH 130
AFN Y D+GLFG + + A ++ + SVT EE ERAK QL + LL +
Sbjct 348 AFNHSYTDSGLFGMFVTILDDAAHLAAPLIIRELCNTVLSVTSEETERAKNQLKSSLLMN 407
Query 131 LDSTTSVAEDLGRQILVY-GRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGP 189
L+S EDLGRQI G ++ E + +++A+ ++ RVA + L VS G
Sbjct 408 LESRMISLEDLGRQIQTQNGLYITPKEMIEKIDALTPSDLSRVARRVLTGN---VSNPGN 464
Query 190 LHGMPQLI 197
G P ++
Sbjct 465 GTGKPTVL 472
> 7294089
Length=440
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 50/175 (28%), Positives = 80/175 (45%), Gaps = 14/175 (8%)
Query 22 DVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRDTGL 81
+ +AF +++ +G+ + G G A G A N Y D GL
Sbjct 272 EALAFAILEQALGAKAATKRGTSAGLFGE-------AVNCAGGVGASVKAVNASYSDAGL 324
Query 82 FGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDL 141
FGF D + V L+ G+ S SV+D++V R KA L +++ S + +++
Sbjct 325 FGFVVSADSKDIGKTVEFLVRGLK--SASVSDKDVARGKALLKARIISRYSSDGGLIKEI 382
Query 142 GRQILVYGRHVPTAEFLMRLNAIDADEIKRV--AWKYLHDKEVAVSAMGPLHGMP 194
GRQ + R+V A+ L L AID +V A K + ++AV A+G L +P
Sbjct 383 GRQAALT-RNVLEADAL--LGAIDGISQSQVQEAAKKVGSSKLAVGAIGHLANVP 434
> At1g51980
Length=503
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 62/129 (48%), Gaps = 8/129 (6%)
Query 70 SAFNTCYRDTGLFGFYA----QCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTT 125
+AF + + DTGLFG Y Q A+E +L V ++RAKA +
Sbjct 369 TAFTSIFNDTGLFGIYGCSSPQFAAKAIELAAKELKD---VAGGKVNQAHLDRAKAATKS 425
Query 126 QLLGHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVS 185
+L +L+S AED+GRQIL YG P +FL ++ + +I K + K + +
Sbjct 426 AVLMNLESRMIAAEDIGRQILTYGERKPVDQFLKSVDQLTLKDIADFTSKVI-SKPLTMG 484
Query 186 AMGPLHGMP 194
+ G + +P
Sbjct 485 SFGDVLAVP 493
> At3g16480
Length=448
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 61/126 (48%), Gaps = 2/126 (1%)
Query 70 SAFNTCYRDTGLFGFYA-QCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLL 128
+AF + + +TGLFG Y E A + L V + ++RAKA + +L
Sbjct 314 TAFTSVFNNTGLFGIYGCTSPEFASQGIELVASEMNAVADGKVNQKHLDRAKAATKSAIL 373
Query 129 GHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L+S AED+GRQIL YG P +FL ++ + +I K + K + ++ G
Sbjct 374 MNLESRMIAAEDIGRQILTYGERKPVDQFLKTVDQLTLKDIADFTSKVI-TKPLTMATFG 432
Query 189 PLHGMP 194
+ +P
Sbjct 433 DVLNVP 438
> CE11226
Length=471
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 61/127 (48%), Gaps = 3/127 (2%)
Query 72 FNTCYRDTGLFGFYAQCDEVAVEHC---VLDLMHGITALSYSVTDEEVERAKAQLTTQLL 128
FN Y+DTGLFG Y D + + + H L+ + T+EEV AK Q T L
Sbjct 332 FNINYKDTGLFGIYFVADAHDLNDTSGIMKSVAHEWKHLASAATEEEVAMAKNQFRTNLY 391
Query 129 GHLDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L++ T A +++L G +E ++ +DA ++ ++++D+++A +G
Sbjct 392 QNLETNTQKAGFNAKELLYTGNLRQLSELEAQIQKVDAGAVREAISRHVYDRDLAAVGVG 451
Query 189 PLHGMPQ 195
P
Sbjct 452 RTEAFPN 458
> Hs20535969
Length=417
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 63/126 (50%), Gaps = 3/126 (2%)
Query 70 SAFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLG 129
+++ Y DTGL A D V V + +V E+ERAK QLT+ L+
Sbjct 270 TSYYLSYEDTGLLCILASADPRQVPEMVEITTKEFILMGGTVDAVELERAKTQLTSMLMM 329
Query 130 HLDSTTSVAEDLGRQIL-VYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMG 188
+L+S + ED+GRQ+L R +P E + +++KRVA K L K V V+A+G
Sbjct 330 NLESRPVIFEDVGRQVLATRSRKLP-QELCTFIRNEKPEDVKRVASKILQGKPV-VAALG 387
Query 189 PLHGMP 194
+P
Sbjct 388 KSTDLP 393
> Hs4507843
Length=453
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 43/178 (24%), Positives = 89/178 (50%), Gaps = 12/178 (6%)
Query 20 CADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMF--SAFNTCYR 77
A+ AF ++Q ++G+ + G +NTT ++ + + F SAFN Y
Sbjct 280 SAEANAFSVLQHVLGAGPHVKRG-------SNTT-SHLHQAVAKATQQPFDVSAFNASYS 331
Query 78 DTGLFGFYAQCDEVAVEHCVLDLMHGITALSY-SVTDEEVERAKAQLTTQLLGHLDSTTS 136
D+GLFG Y A + + + ++ ++++ +V+ AK +L L ++S+
Sbjct 332 DSGLFGIYTISQATAAGDVIKAAYNQVKRIAQGNLSNTDVQAAKNKLKAGYLMSVESSEC 391
Query 137 VAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAVSAMGPLHGMP 194
E++G Q LV G ++P + L +++++ +I A K++ ++ +++A G L P
Sbjct 392 FLEEVGSQALVAGSYMPPSTVLQQIDSVANADIINAAKKFVSGQK-SMAASGNLGHTP 448
> CE27028
Length=477
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 53/110 (48%), Gaps = 0/110 (0%)
Query 71 AFNTCYRDTGLFGFYAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGH 130
A N Y D+G+F A + ++ L+H I L V E+ RA+ QL + L+ +
Sbjct 336 AHNHSYSDSGVFTVTASSPPENINDALILLVHQILQLQQGVEPTELARARTQLRSHLMMN 395
Query 131 LDSTTSVAEDLGRQILVYGRHVPTAEFLMRLNAIDADEIKRVAWKYLHDK 180
L+ + ED+ RQ+L +G E+ ++ + +I RV + L K
Sbjct 396 LEVRPVLFEDMVRQVLGHGDRKQPEEYAEKIEKVTNSDIIRVTERLLASK 445
> Hs4885069
Length=244
Score = 35.8 bits (81), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 34/62 (54%), Gaps = 3/62 (4%)
Query 85 YAQCDEVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDST---TSVAEDL 141
Y +C + E+ V D+ H T + T++ V+R K+Q T+ + H+ S ++VA DL
Sbjct 174 YIECSALQSENSVRDIFHVATLACVNKTNKNVKRNKSQRATKRISHMPSRPELSAVATDL 233
Query 142 GR 143
+
Sbjct 234 RK 235
> Hs20270311
Length=418
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 5/92 (5%)
Query 19 LCADVVAFMLMQAIIGSYRKHEEGLVPGKLSANTTVRNIATKMQTGCAEMFSAFNTCYRD 78
LC+DV+ + L + +R H +G + NT + + T M NT ++
Sbjct 325 LCSDVILYPLETVL---HRLHIQGTR--TIIDNTDLGYEVLPINTQYEGMRDCINTIRQE 379
Query 79 TGLFGFYAQCDEVAVEHCVLDLMHGITALSYS 110
G+FGFY V +++ + + IT + YS
Sbjct 380 EGVFGFYKGFGAVIIQYTLHAAVLQITKIIYS 411
> Hs7662474
Length=1709
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 28/90 (31%), Positives = 44/90 (48%), Gaps = 12/90 (13%)
Query 87 QCDEVAVEHCV---LDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDLGR 143
+C + +EH L +HG TAL Y+V +E++ A T LL + +DL
Sbjct 127 KCATILLEHGADPNLADVHGNTALHYAVYNEDISVA----TKLLLYDANIEAKNKDDLTP 182
Query 144 QIL-VYGRHVPTAEFLMR----LNAIDADE 168
+L V G+ EFL++ +NA+D E
Sbjct 183 LLLAVSGKKQQMVEFLIKKKANVNAVDKLE 212
> 7300730
Length=319
Score = 28.5 bits (62), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 48/96 (50%), Gaps = 14/96 (14%)
Query 90 EVAVEHCVLDLMHGITALSYSVTDEEVERAKAQLTTQLLGHLDSTTSVAEDLG-RQILVY 148
E+ ++HC +L+ + + ++T+++ R LDS A D G ++++V+
Sbjct 25 ELDMKHCFENLI--VIDVGANLTNKKYSR-----------DLDSVVQRARDAGVQKLMVH 71
Query 149 GRHVPTAEFLMRLNAIDADEIKRVAWKYLHDKEVAV 184
G V +++ +RL+ I D I A + HD + V
Sbjct 72 GTSVKSSKEALRLSRIYPDIIYSTAGIHPHDSKSIV 107
Lambda K H
0.323 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3371754244
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40