bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3620_orf2
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
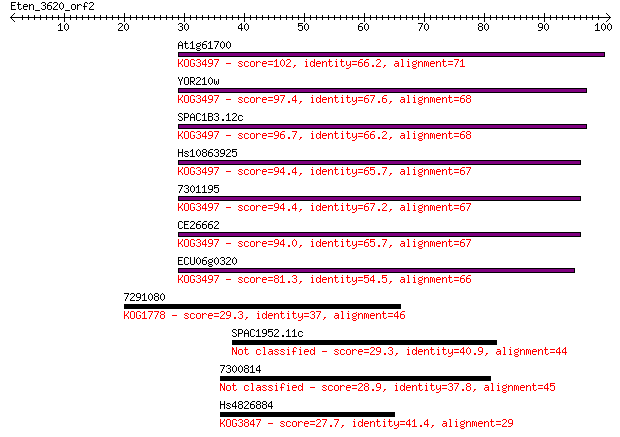
At1g61700 102 2e-22
YOR210w 97.4 5e-21
SPAC1B3.12c 96.7 8e-21
Hs10863925 94.4 5e-20
7301195 94.4 5e-20
CE26662 94.0 7e-20
ECU06g0320 81.3 4e-16
7291080 29.3 1.6
SPAC1952.11c 29.3 1.8
7300814 28.9 2.6
Hs4826884 27.7 5.0
> At1g61700
Length=71
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 47/71 (66%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 29 MIIPVRCFTCGRVIGHLWEQYVEKLQNDVDEGVALDDLGLKRYCCRRMILTHADLIDKLL 88
MI+PVRCFTCG+VIG+ W+ Y+E LQ D EG ALD LGL RYCCRRM++TH DLI+KLL
Sbjct 1 MIVPVRCFTCGKVIGNKWDTYLELLQADYAEGDALDALGLVRYCCRRMLMTHVDLIEKLL 60
Query 89 AYNIYEKKLPD 99
YN EK P+
Sbjct 61 NYNTMEKSDPN 71
> YOR210w
Length=70
Score = 97.4 bits (241), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 46/69 (66%), Positives = 54/69 (78%), Gaps = 1/69 (1%)
Query 29 MIIPVRCFTCGRVIGHLWEQYVEKLQND-VDEGVALDDLGLKRYCCRRMILTHADLIDKL 87
MI+PVRCF+CG+V+G WE Y+ LQ D +DEG AL LGLKRYCCRRMILTH DLI+K
Sbjct 1 MIVPVRCFSCGKVVGDKWESYLNLLQEDELDEGTALSRLGLKRYCCRRMILTHVDLIEKF 60
Query 88 LAYNIYEKK 96
L YN EK+
Sbjct 61 LRYNPLEKR 69
> SPAC1B3.12c
Length=71
Score = 96.7 bits (239), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 45/68 (66%), Positives = 53/68 (77%), Gaps = 0/68 (0%)
Query 29 MIIPVRCFTCGRVIGHLWEQYVEKLQNDVDEGVALDDLGLKRYCCRRMILTHADLIDKLL 88
MIIP+RCF+CG+VIG W+ Y+ LQ D EG ALD LGL+RYCCRRMILTH DLI+KLL
Sbjct 1 MIIPIRCFSCGKVIGDKWDTYLTLLQEDNTEGEALDKLGLQRYCCRRMILTHVDLIEKLL 60
Query 89 AYNIYEKK 96
YN K+
Sbjct 61 CYNPLSKQ 68
> Hs10863925
Length=67
Score = 94.4 bits (233), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 44/67 (65%), Positives = 52/67 (77%), Gaps = 0/67 (0%)
Query 29 MIIPVRCFTCGRVIGHLWEQYVEKLQNDVDEGVALDDLGLKRYCCRRMILTHADLIDKLL 88
MIIPVRCFTCG+++G+ WE Y+ LQ + EG ALD LGLKRYCCRRM+L H DLI+KLL
Sbjct 1 MIIPVRCFTCGKIVGNKWEAYLGLLQAEYTEGDALDALGLKRYCCRRMLLAHVDLIEKLL 60
Query 89 AYNIYEK 95
Y EK
Sbjct 61 NYAPLEK 67
> 7301195
Length=67
Score = 94.4 bits (233), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 45/67 (67%), Positives = 52/67 (77%), Gaps = 0/67 (0%)
Query 29 MIIPVRCFTCGRVIGHLWEQYVEKLQNDVDEGVALDDLGLKRYCCRRMILTHADLIDKLL 88
MIIP+RCFTCG+VIG+ WE Y+ LQ + EG ALD LGLKRYCCRRM+L H DLI+KLL
Sbjct 1 MIIPIRCFTCGKVIGNKWESYLGLLQAEYTEGDALDALGLKRYCCRRMLLGHVDLIEKLL 60
Query 89 AYNIYEK 95
Y EK
Sbjct 61 NYAPLEK 67
> CE26662
Length=67
Score = 94.0 bits (232), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 44/67 (65%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 29 MIIPVRCFTCGRVIGHLWEQYVEKLQNDVDEGVALDDLGLKRYCCRRMILTHADLIDKLL 88
MIIP+RCFTCG+VIG WE Y+ LQ++ EG ALD LGL+RYCCRRM+L H DLI+KLL
Sbjct 1 MIIPIRCFTCGKVIGDKWETYLGFLQSEYSEGDALDALGLRRYCCRRMLLAHVDLIEKLL 60
Query 89 AYNIYEK 95
Y+ EK
Sbjct 61 NYHPLEK 67
> ECU06g0320
Length=72
Score = 81.3 bits (199), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 36/66 (54%), Positives = 48/66 (72%), Gaps = 0/66 (0%)
Query 29 MIIPVRCFTCGRVIGHLWEQYVEKLQNDVDEGVALDDLGLKRYCCRRMILTHADLIDKLL 88
MIIP+RCFTCG+ I WE Y E L+ D + ALD+LG++R CCRRM L H D++DKLL
Sbjct 1 MIIPIRCFTCGKEISSKWEPYQELLKIDKGKAEALDELGMRRICCRRMFLGHVDIVDKLL 60
Query 89 AYNIYE 94
++I +
Sbjct 61 KFDIVK 66
> 7291080
Length=3275
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 25/57 (43%), Gaps = 13/57 (22%)
Query 20 FLFILFFCKMIIPVR-----------CFTCGRVIGHLWEQYVEKLQNDVDEGVALDD 65
F++ CK + R C C +GH + +EKL D+D+G AL D
Sbjct 2330 FVYTCNHCKTAVETRYHCTVCDDFDLCIVCKEKVGH--QHKMEKLGFDIDDGSALAD 2384
> SPAC1952.11c
Length=835
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 7/51 (13%)
Query 38 CGRVIGHLWEQ-YVEKLQNDV---DEGVALDDLGLKRYCCRRMI---LTHA 81
CG VI W+ + KLQ DEG +D+ +KRY + I +TH
Sbjct 637 CGEVISRTWKTAHKNKLQRGALPEDEGSGVDNFRVKRYVSKYTINPAITHG 687
> 7300814
Length=1483
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 12/57 (21%)
Query 36 FTCGRVIGHLWEQYVEKLQNDVDEGV-----ALDDLGLKRYCCR-------RMILTH 80
C +V W Q VEK++ D++E + ALD+ +R C M+ TH
Sbjct 903 LACAQVEAEEWRQQVEKIRTDLEEQIRILKNALDNSEAERKICEDKWQKEFEMLRTH 959
> Hs4826884
Length=441
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 18/29 (62%), Gaps = 5/29 (17%)
Query 36 FTCGRVIGHLWEQYVEKLQNDVDEGVALD 64
F G++IGH+ KL+ D+D VA+D
Sbjct 359 FATGKIIGHML-----KLKGDIDSNVAID 382
Lambda K H
0.333 0.151 0.518
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187882580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40