bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3606_orf1
Length=247
Score E
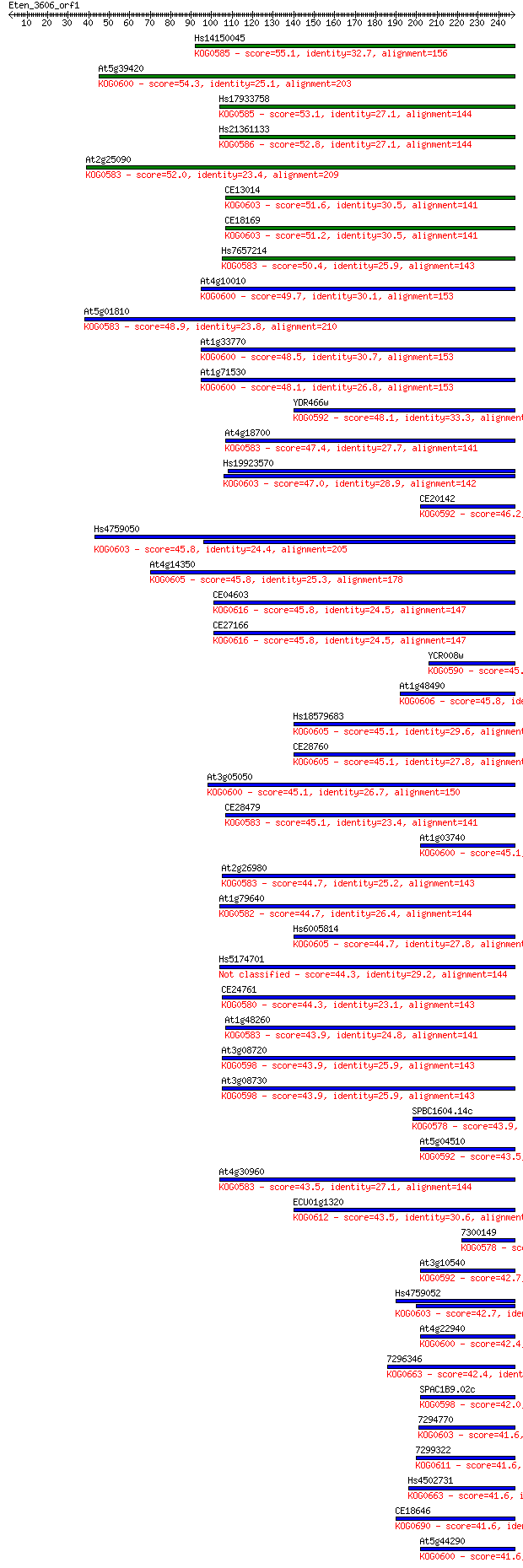
Sequences producing significant alignments: (Bits) Value
Hs14150045 55.1 1e-07
At5g39420 54.3 2e-07
Hs17933758 53.1 6e-07
Hs21361133 52.8 7e-07
At2g25090 52.0 1e-06
CE13014 51.6 2e-06
CE18169 51.2 2e-06
Hs7657214 50.4 3e-06
At4g10010 49.7 5e-06
At5g01810 48.9 9e-06
At1g33770 48.5 1e-05
At1g71530 48.1 2e-05
YDR466w 48.1 2e-05
At4g18700 47.4 3e-05
Hs19923570 47.0 3e-05
CE20142 46.2 6e-05
Hs4759050 45.8 8e-05
At4g14350 45.8 8e-05
CE04603 45.8 8e-05
CE27166 45.8 9e-05
YCR008w 45.8 9e-05
At1g48490 45.8 9e-05
Hs18579683 45.1 1e-04
CE28760 45.1 1e-04
At3g05050 45.1 1e-04
CE28479 45.1 1e-04
At1g03740 45.1 2e-04
At2g26980 44.7 2e-04
At1g79640 44.7 2e-04
Hs6005814 44.7 2e-04
Hs5174701 44.3 2e-04
CE24761 44.3 3e-04
At1g48260 43.9 3e-04
At3g08720 43.9 3e-04
At3g08730 43.9 3e-04
SPBC1604.14c 43.9 3e-04
At5g04510 43.5 4e-04
At4g30960 43.5 4e-04
ECU01g1320 43.5 4e-04
7300149 42.7 6e-04
At3g10540 42.7 6e-04
Hs4759052 42.7 6e-04
At4g22940 42.4 9e-04
7296346 42.4 0.001
SPAC1B9.02c 42.0 0.001
7294770 41.6 0.001
7299322 41.6 0.001
Hs4502731 41.6 0.001
CE18646 41.6 0.001
At5g44290 41.6 0.001
> Hs14150045
Length=505
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 51/167 (30%), Positives = 70/167 (41%), Gaps = 55/167 (32%)
Query 92 RPGPTG----MGFMAK----MEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFI 143
RP P G G AK +E V +E IL LDH +KL +VLD + A+ +Y +
Sbjct 173 RPPPRGSQAAQGGPAKQLLPLERVYQEIAILKKLDHVNVVKLIEVLD---DPAEDNLYLV 229
Query 144 TNFLPGGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFT 203
+ L G +M ++ D P F+
Sbjct 230 FDLLRKGPVM--------------EVPCDKP---------------------------FS 248
Query 204 EAQAKCFIRDAAEVHEHL---GICHRDLKVDNLLLGADGRVCVGDFG 247
E QA+ ++RD E+L I HRD+K NLLLG DG V + DFG
Sbjct 249 EEQARLYLRDVILGLEYLHCQKIVHRDIKPSNLLLGDDGHVKIADFG 295
> At5g39420
Length=576
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 51/207 (24%), Positives = 82/207 (39%), Gaps = 66/207 (31%)
Query 45 YRVCEKLASTELSSVFRVEEANPASGVTTSFCCKRYRKMLLLRRREY-RPGPTGMGFMAK 103
++ EK+ SSVFR E KM+ L++ ++ P + FMA+
Sbjct 105 FQKLEKIGQGTYSSVFRAREVETG-------------KMVALKKVKFDNLQPESIRFMAR 151
Query 104 MEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEEL 163
E IL L+HP +KL+ ++ S S+ +Y + ++
Sbjct 152 ------EILILRKLNHPNIMKLEGIVTSRASSS---IYLVFEYM---------------- 186
Query 164 VRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEH--- 220
E +A +S DI FTE Q KC+++ EH
Sbjct 187 -----------EHDLAGLSSNPDI-------------RFTEPQIKCYMKQLLWGLEHCHM 222
Query 221 LGICHRDLKVDNLLLGADGRVCVGDFG 247
G+ HRD+K N+L+ G + +GDFG
Sbjct 223 RGVIHRDIKASNILVNNKGVLKLGDFG 249
> Hs17933758
Length=588
Score = 53.1 bits (126), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 39/147 (26%), Positives = 62/147 (42%), Gaps = 47/147 (31%)
Query 104 MEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEEL 163
+E V +E IL LDHP +KL +VLD + + +Y + + G +M
Sbjct 230 IEQVYQEIAILKKLDHPNVVKLVEVLD---DPNEDHLYMVFELVNQGPVM---------- 276
Query 164 VRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL-- 221
++ TL+ +E QA+ + +D + E+L
Sbjct 277 -------------------------------EVPTLKPLSEDQARFYFQDLIKGIEYLHY 305
Query 222 -GICHRDLKVDNLLLGADGRVCVGDFG 247
I HRD+K NLL+G DG + + DFG
Sbjct 306 QKIIHRDIKPSNLLVGEDGHIKIADFG 332
> Hs21361133
Length=752
Score = 52.8 bits (125), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 39/148 (26%), Positives = 67/148 (45%), Gaps = 27/148 (18%)
Query 104 MEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGA----LMVLNPMR 159
++ + E RI+ +L+HP +KL +V+++ + +Y I + GG L+ M+
Sbjct 97 LQKLFREVRIMKILNHPNIVKLFEVIET-----EKTLYLIMEYASGGEVFDYLVAHGKMK 151
Query 160 DEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHE 219
++E + A + + +S D++ FT FI A +
Sbjct 152 EKEARSKFRQGCQAGQTIKVQVSF--DLLSLMFT----------------FIVSAVQYCH 193
Query 220 HLGICHRDLKVDNLLLGADGRVCVGDFG 247
I HRDLK +NLLL AD + + DFG
Sbjct 194 QKRIVHRDLKAENLLLDADMNIKIADFG 221
> At2g25090
Length=469
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 49/209 (23%), Positives = 87/209 (41%), Gaps = 58/209 (27%)
Query 39 SVWLNQYRVCEKLASTELSSVFRVEEANPASGVTTSFCCKRYRKMLLLRRREYRPGPTGM 98
+V ++Y + L + + V+ E + V K +K + +RR GM
Sbjct 9 TVLFDKYNIGRLLGTGNFAKVYHGTEISTGDDV----AIKVIKKDHVFKRR-------GM 57
Query 99 GFMAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPM 158
ME ++ E ++ LL HP ++L++V+ + K++F+ ++ GG L
Sbjct 58 -----MEQIEREIAVMRLLRHPNVVELREVM-----ATKKKIFFVMEYVNGGELF----- 102
Query 159 RDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVH 218
E + D + ED+ R K+ QL + F ++
Sbjct 103 ----------------EMIDRDGKLPEDLAR-KYFQQLISAVDFCHSR------------ 133
Query 219 EHLGICHRDLKVDNLLLGADGRVCVGDFG 247
G+ HRD+K +NLLL +G + V DFG
Sbjct 134 ---GVFHRDIKPENLLLDGEGDLKVTDFG 159
> CE13014
Length=740
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 58/144 (40%), Gaps = 48/144 (33%)
Query 107 VKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELVRS 166
K E ILA + HP +KL + +GK+Y I +FL GG L
Sbjct 94 TKLERNILAHISHPFIVKLHYAFQT-----EGKLYLILDFLRGGDLF------------- 135
Query 167 CQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL---GI 223
+LS + FTE K ++ + EHL GI
Sbjct 136 TRLSKEV---------------------------MFTEDDVKFYLAELTLALEHLHSLGI 168
Query 224 CHRDLKVDNLLLGADGRVCVGDFG 247
+RDLK +N+LL ADG + V DFG
Sbjct 169 VYRDLKPENILLDADGHIKVTDFG 192
> CE18169
Length=797
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 58/144 (40%), Gaps = 48/144 (33%)
Query 107 VKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELVRS 166
K E ILA + HP +KL + +GK+Y I +FL GG L
Sbjct 151 TKLERNILAHISHPFIVKLHYAFQT-----EGKLYLILDFLRGGDLF------------- 192
Query 167 CQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL---GI 223
+LS + FTE K ++ + EHL GI
Sbjct 193 TRLSKEV---------------------------MFTEDDVKFYLAELTLALEHLHSLGI 225
Query 224 CHRDLKVDNLLLGADGRVCVGDFG 247
+RDLK +N+LL ADG + V DFG
Sbjct 226 VYRDLKPENILLDADGHIKVTDFG 249
> Hs7657214
Length=714
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 60/146 (41%), Gaps = 48/146 (32%)
Query 105 EDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELV 164
++++ E +I ++ HP +L D+L++ + Y + PGG LM
Sbjct 106 KNLRREGQIQQMIRHPNITQLLDILET-----ENSYYLVMELCPGGNLM----------- 149
Query 165 RSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL--- 221
I EK + E++A+ +IR EHL
Sbjct 150 ---------------------HKIYEK--------KRLEESEARRYIRQLISAVEHLHRA 180
Query 222 GICHRDLKVDNLLLGADGRVCVGDFG 247
G+ HRDLK++NLLL D + + DFG
Sbjct 181 GVVHRDLKIENLLLDEDNNIKLIDFG 206
> At4g10010
Length=649
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 46/156 (29%), Positives = 62/156 (39%), Gaps = 52/156 (33%)
Query 95 PTGMGFMAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMV 154
P + FMA+ E IL LDHP +KL+ +T+ L G +V
Sbjct 194 PESVRFMAR------EINILRKLDHPNVMKLE--------------CLVTSKLSGSLYLV 233
Query 155 LNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDA 214
M + LS A P V FTE+Q KC+++
Sbjct 234 FEYMEHD-------LSGLALRPGVK----------------------FTESQIKCYMKQL 264
Query 215 AEVHEHL---GICHRDLKVDNLLLGADGRVCVGDFG 247
EH GI HRD+K NLL+ DG + +GDFG
Sbjct 265 LSGLEHCHSRGILHRDIKGPNLLVNNDGVLKIGDFG 300
> At5g01810
Length=421
Score = 48.9 bits (115), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 50/210 (23%), Positives = 74/210 (35%), Gaps = 60/210 (28%)
Query 38 GSVWLNQYRVCEKLASTELSSVFRVEEANPASGVTTSFCCKRYRKMLLLRRREYRPGPTG 97
GSV + +Y V + L + V+ V K R G
Sbjct 5 GSVLMLRYEVGKFLGQGTFAKVYHARHLKTGDSVAIKVIDKE------------RILKVG 52
Query 98 MGFMAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNP 157
M E +K E + LL HP ++L +V+ + K+YF+ + GG L
Sbjct 53 M-----TEQIKREISAMRLLRHPNIVELHEVM-----ATKSKIYFVMEHVKGGELF---- 98
Query 158 MRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEV 217
V+ + ED+ R+ F + A C R
Sbjct 99 ------------------NKVSTGKLREDVARKYF-------QQLVRAVDFCHSR----- 128
Query 218 HEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
G+CHRDLK +NLLL G + + DFG
Sbjct 129 ----GVCHRDLKPENLLLDEHGNLKISDFG 154
> At1g33770
Length=614
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 47/156 (30%), Positives = 61/156 (39%), Gaps = 52/156 (33%)
Query 95 PTGMGFMAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMV 154
P + FMA+ E IL LDHP +KL+ +T+ L G +V
Sbjct 179 PESVRFMAR------EINILRKLDHPNVMKLQ--------------CLVTSKLSGSLHLV 218
Query 155 LNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDA 214
M + LS A P V FTE Q KCF++
Sbjct 219 FEYMEHD-------LSGLALRPGV----------------------KFTEPQIKCFMKQL 249
Query 215 AEVHEHL---GICHRDLKVDNLLLGADGRVCVGDFG 247
EH GI HRD+K NLL+ DG + +GDFG
Sbjct 250 LCGLEHCHSRGILHRDIKGSNLLVNNDGVLKIGDFG 285
> At1g71530
Length=655
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 41/156 (26%), Positives = 63/156 (40%), Gaps = 52/156 (33%)
Query 95 PTGMGFMAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMV 154
P + FMA+ E IL LDHP +KL+ ++ S G +Y + ++
Sbjct 185 PESVRFMAR------EILILRKLDHPNVMKLEGLVTSR---LSGSLYLVFEYM------- 228
Query 155 LNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDA 214
E +A ++ T I F+E Q KC+++
Sbjct 229 --------------------EHDLAGLAATPGI-------------KFSEPQIKCYMQQL 255
Query 215 AEVHEH---LGICHRDLKVDNLLLGADGRVCVGDFG 247
EH GI HRD+K NLL+ +G + +GDFG
Sbjct 256 FRGLEHCHRRGILHRDIKGSNLLINNEGVLKIGDFG 291
> YDR466w
Length=898
Score = 48.1 bits (113), Expect = 2e-05, Method: Composition-based stats.
Identities = 36/109 (33%), Positives = 46/109 (42%), Gaps = 39/109 (35%)
Query 140 VYFITNFLPGGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETL 199
+YF+ +F PGG L+ L + M DI FT QL
Sbjct 88 LYFVLDFAPGGELLSL----------------------LHKMGTFNDIWTRHFTAQL--- 122
Query 200 RTFTEAQAKCFIRDAAE-VHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
DA E +H H GI HRDLK +N+LL DGR+ + DFG
Sbjct 123 ------------IDALEFIHSH-GIIHRDLKPENVLLDRDGRLMITDFG 158
> At4g18700
Length=489
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 60/141 (42%), Gaps = 43/141 (30%)
Query 107 VKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELVRS 166
+K E IL + HP ++L +V+ + K+YF+ ++ GG L
Sbjct 71 IKREISILRRVRHPNIVQLFEVM-----ATKAKIYFVMEYVRGGELF------------- 112
Query 167 CQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLGICHR 226
VA + E++ R K+ QL + TF A+ G+ HR
Sbjct 113 ---------NKVAKGRLKEEVAR-KYFQQLISAVTFCHAR---------------GVYHR 147
Query 227 DLKVDNLLLGADGRVCVGDFG 247
DLK +NLLL +G + V DFG
Sbjct 148 DLKPENLLLDENGNLKVSDFG 168
> Hs19923570
Length=733
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 40/143 (27%), Positives = 59/143 (41%), Gaps = 48/143 (33%)
Query 108 KEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELVRSC 167
K E ILA ++HP +KL + +GK+Y I +FL GG L
Sbjct 107 KMERDILAEVNHPFIVKLHYAFQT-----EGKLYLILDFLRGGDLF-------------T 148
Query 168 QLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL---GIC 224
+LS + FTE K ++ + A +HL GI
Sbjct 149 RLSKEV---------------------------MFTEEDVKFYLAELALALDHLHSLGII 181
Query 225 HRDLKVDNLLLGADGRVCVGDFG 247
+RDLK +N+LL +G + + DFG
Sbjct 182 YRDLKPENILLDEEGHIKITDFG 204
Score = 33.5 bits (75), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 40/151 (26%), Positives = 56/151 (37%), Gaps = 55/151 (36%)
Query 106 DVKEEARILALL-DHPRCLKLKDVLDSNPESADGK-VYFITNFLPGGALMVLNPMRDEEL 163
D EE IL HP + LKDV D DGK VY + + GG L+
Sbjct 452 DPSEEIEILLRYGQHPNIITLKDVYD------DGKFVYLVMELMRGGELL---------- 495
Query 164 VRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAK---CFIRDAAEVHEH 220
+ I+R+++ F+E +A C I +
Sbjct 496 ---------------------DRILRQRY---------FSEREASDVLCTITKTMDYLHS 525
Query 221 LGICHRDLKVDNLLL----GADGRVCVGDFG 247
G+ HRDLK N+L G+ + V DFG
Sbjct 526 QGVVHRDLKPSNILYRDESGSPESIRVCDFG 556
> CE20142
Length=320
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 30/49 (61%), Gaps = 3/49 (6%)
Query 202 FTEAQAKCFIRDAAEVHEH---LGICHRDLKVDNLLLGADGRVCVGDFG 247
F A ++ + + EH LGI HRD+K DNLL+ +DGR+ + DFG
Sbjct 132 FNVADSRYYAANLLSALEHIHKLGIIHRDVKADNLLVKSDGRIMLTDFG 180
> Hs4759050
Length=740
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 50/208 (24%), Positives = 78/208 (37%), Gaps = 63/208 (30%)
Query 43 NQYRVCEKLASTELSSVFRVEEANPASGVTTSFCCKRYRKMLLLRRREYRPGPTGMGFMA 102
+Q+ + + L VF V++ + S + K +K L R R
Sbjct 66 SQFELLKVLGQGSFGKVFLVKKIS-GSDARQLYAMKVLKKATLKVRDRVR---------- 114
Query 103 KMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEE 162
K E IL ++HP +KL + +GK+Y I +FL GG L
Sbjct 115 ----TKMERDILVEVNHPFIVKLHYAFQT-----EGKLYLILDFLRGGDLF--------- 156
Query 163 LVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL- 221
+LS + FTE K ++ + A +HL
Sbjct 157 ----TRLSKEV---------------------------MFTEEDVKFYLAELALALDHLH 185
Query 222 --GICHRDLKVDNLLLGADGRVCVGDFG 247
GI +RDLK +N+LL +G + + DFG
Sbjct 186 SLGIIYRDLKPENILLDEEGHIKLTDFG 213
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 46/166 (27%), Positives = 63/166 (37%), Gaps = 60/166 (36%)
Query 96 TGMGFMAKM-----EDVKEEARILALL-DHPRCLKLKDVLDSNPESADGK-VYFITNFLP 148
T M F K+ D EE IL HP + LKDV D DGK VY +T +
Sbjct 444 TNMEFAVKIIDKSKRDPTEEIEILLRYGQHPNIITLKDVYD------DGKYVYVVTELMK 497
Query 149 GGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAK 208
GG L+ + I+R+KF F+E +A
Sbjct 498 GGELL-------------------------------DKILRQKF---------FSEREAS 517
Query 209 CFIRDAAEVHEHL---GICHRDLKVDNLLL----GADGRVCVGDFG 247
+ + E+L G+ HRDLK N+L G + + DFG
Sbjct 518 AVLFTITKTVEYLHAQGVVHRDLKPSNILYVDESGNPESIRICDFG 563
> At4g14350
Length=475
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 45/182 (24%), Positives = 68/182 (37%), Gaps = 63/182 (34%)
Query 70 GVTTSFCCKRYRKMLLLRRREYRPGPTGMGFMAKMEDVKEEARILALLDHPRCLKLKDVL 129
G + K+ +K +LRR ++E VK E +LA +D +KL
Sbjct 64 GTGNVYAMKKLKKSEMLRR-------------GQVEHVKAERNLLAEVDSNCIVKLYCSF 110
Query 130 DSNPESADGKVYFITNFLPGGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIR 189
+ +Y I +LPGG +M L +D
Sbjct 111 QD-----EEYLYLIMEYLPGGDMMTLLMRKD----------------------------- 136
Query 190 EKFTPQLETLRTFTEAQAKCFIRDAA----EVHEHLGICHRDLKVDNLLLGADGRVCVGD 245
T TE +A+ +I + +H+H HRD+K DNLLL DG + + D
Sbjct 137 -----------TLTEDEARFYIGETVLAIESIHKH-NYIHRDIKPDNLLLDKDGHMKLSD 184
Query 246 FG 247
FG
Sbjct 185 FG 186
> CE04603
Length=371
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 56/150 (37%), Gaps = 48/150 (32%)
Query 101 MAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRD 160
M + + V E R+L L HP +K+ E +Y I F+PGG +
Sbjct 102 MRQTQHVHNEKRVLLQLKHPFIVKM-----YASEKDSNHLYMIMEFVPGGEMF------- 149
Query 161 EELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRD---AAEV 217
L R+F+ + A+ + + A E
Sbjct 150 ---------------------------------SYLRASRSFSNSMARFYASEIVCALEY 176
Query 218 HEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
LGI +RDLK +NL+L +G + + DFG
Sbjct 177 IHSLGIVYRDLKPENLMLSKEGHIKMADFG 206
> CE27166
Length=270
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 56/150 (37%), Gaps = 48/150 (32%)
Query 101 MAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRD 160
M + + V E R+L L HP +K+ E +Y I F+PGG +
Sbjct 1 MRQTQHVHNEKRVLLQLKHPFIVKM-----YASEKDSNHLYMIMEFVPGGEMF------- 48
Query 161 EELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRD---AAEV 217
L R+F+ + A+ + + A E
Sbjct 49 ---------------------------------SYLRASRSFSNSMARFYASEIVCALEY 75
Query 218 HEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
LGI +RDLK +NL+L +G + + DFG
Sbjct 76 IHSLGIVYRDLKPENLMLSKEGHIKMADFG 105
> YCR008w
Length=603
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 30/46 (65%), Gaps = 5/46 (10%)
Query 206 QAKCF----IRDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
+A CF IR +HE +G+CHRDLK +NLLL DG + + DFG
Sbjct 425 EADCFFKQLIRGVVYMHE-MGVCHRDLKPENLLLTHDGVLKITDFG 469
> At1g48490
Length=1244
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 32/59 (54%), Gaps = 3/59 (5%)
Query 192 FTPQLETLRTFTEAQAKCFIRDAAEVHEHL---GICHRDLKVDNLLLGADGRVCVGDFG 247
F L + EA A+ +I + E+L G+ HRDLK DNLL+ DG V + DFG
Sbjct 913 FYSMLRKIGCLDEANARVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHVKLTDFG 971
> Hs18579683
Length=464
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 42/111 (37%), Gaps = 43/111 (38%)
Query 140 VYFITNFLPGGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETL 199
+Y I FLPGG +M L +D
Sbjct 163 LYLIMEFLPGGDMMTLLMKKD--------------------------------------- 183
Query 200 RTFTEAQAKCFIRD---AAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
T TE + + +I + A + LG HRD+K DNLLL A G V + DFG
Sbjct 184 -TLTEEETQFYISETVLAIDAIHQLGFIHRDIKPDNLLLDAKGHVKLSDFG 233
> CE28760
Length=467
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 45/108 (41%), Gaps = 37/108 (34%)
Query 140 VYFITNFLPGGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETL 199
+Y + FLPGG +M L L++ L+ +A + +A+ ++
Sbjct 153 LYLVMEFLPGGDMMTL-------LIKKDTLTEEATQFYIAEAAL---------------- 189
Query 200 RTFTEAQAKCFIRDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
A + LG HRD+K DNLLL A G V + DFG
Sbjct 190 --------------AIQFIHSLGFIHRDIKPDNLLLDARGHVKLSDFG 223
> At3g05050
Length=593
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 40/153 (26%), Positives = 64/153 (41%), Gaps = 52/153 (33%)
Query 98 MGFMAKMEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNP 157
+ FMA+ E IL LDHP +KL+ ++ S S+ +Y + ++
Sbjct 179 LKFMAR------EILILRRLDHPNVIKLEGLVTSRMSSS---LYLVFRYM---------- 219
Query 158 MRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEV 217
D +L A ++ + +I FTE Q KC+++
Sbjct 220 --DHDL---------------AGLAASPEI-------------KFTEQQVKCYMKQLLSG 249
Query 218 HEH---LGICHRDLKVDNLLLGADGRVCVGDFG 247
EH G+ HRD+K NLL+ G + +GDFG
Sbjct 250 LEHCHNRGVLHRDIKGSNLLIDDGGVLRIGDFG 282
> CE28479
Length=822
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/141 (23%), Positives = 60/141 (42%), Gaps = 42/141 (29%)
Query 107 VKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELVRS 166
++ E +I+ ++DHP +K +++ D +Y ++ + G L
Sbjct 60 LEREVKIVKVIDHPHIVKSYEIM-----RVDNMLYIVSEYCSSGELY------------- 101
Query 167 CQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLGICHR 226
E ++ V E++ R+ F+ ET A A ++ GI HR
Sbjct 102 --------ETLIEKGRVAENVARKWFS---ET------ASAVAYLHSQ-------GIVHR 137
Query 227 DLKVDNLLLGADGRVCVGDFG 247
DLK +N+LLG + + + DFG
Sbjct 138 DLKAENILLGKNSNIKIIDFG 158
> At1g03740
Length=542
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 29/49 (59%), Gaps = 3/49 (6%)
Query 202 FTEAQAKCFIRDAAEVHEH---LGICHRDLKVDNLLLGADGRVCVGDFG 247
FTE Q KC++R EH G+ HRD+K NLL+ + G + + DFG
Sbjct 309 FTEPQVKCYMRQLLSGLEHCHSRGVLHRDIKGSNLLIDSKGVLKIADFG 357
> At2g26980
Length=441
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 36/143 (25%), Positives = 57/143 (39%), Gaps = 42/143 (29%)
Query 105 EDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELV 164
E ++ E + L+ HP ++L +V+ S K++ I ++ GG L
Sbjct 57 EQIRREIATMKLIKHPNVVQLYEVMASKT-----KIFIILEYVTGGELF----------- 100
Query 165 RSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLGIC 224
+ +V D + ED R F + A C R G+
Sbjct 101 ----------DKIVNDGRMKEDEARRYF-------QQLIHAVDYCHSR---------GVY 134
Query 225 HRDLKVDNLLLGADGRVCVGDFG 247
HRDLK +NLLL + G + + DFG
Sbjct 135 HRDLKPENLLLDSYGNLKISDFG 157
> At1g79640
Length=547
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/144 (26%), Positives = 59/144 (40%), Gaps = 40/144 (27%)
Query 104 MEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEEL 163
+ ++ EA+ + L+DHP LK S D ++ I ++ GG
Sbjct 53 LNNISREAQTMMLVDHPNVLKSHCSFVS-----DHNLWVIMPYMSGG------------- 94
Query 164 VRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLGI 223
SC A P + ++ I+RE ++ +H+H G
Sbjct 95 --SCLHILKAAYPDGFEEAIIATILREA-------------------LKGLDYLHQH-GH 132
Query 224 CHRDLKVDNLLLGADGRVCVGDFG 247
HRD+K N+LLGA G V +GDFG
Sbjct 133 IHRDVKAGNILLGARGAVKLGDFG 156
> Hs6005814
Length=465
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 44/108 (40%), Gaps = 37/108 (34%)
Query 140 VYFITNFLPGGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETL 199
+Y I FLPGG +M L L++ L+ + + +A+ + D I +
Sbjct 162 LYLIMEFLPGGDMMTL-------LMKKDTLTEEETQFYIAETVLAIDSIHQ--------- 205
Query 200 RTFTEAQAKCFIRDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
LG HRD+K DNLLL + G V + DFG
Sbjct 206 ---------------------LGFIHRDIKPDNLLLDSKGHVKLSDFG 232
> Hs5174701
Length=968
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 42/147 (28%), Positives = 54/147 (36%), Gaps = 47/147 (31%)
Query 104 MEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEEL 163
+ED E ILA DHP +KL DGK++ + F PGGA+
Sbjct 75 LEDYIVEIEILATCDHPYIVKLLGAY-----YHDGKLWIMIEFCPGGAV----------- 118
Query 164 VRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLG- 222
D I LE R TE Q + R E L
Sbjct 119 ----------------------DAI------MLELDRGLTEPQIQVVCRQMLEALNFLHS 150
Query 223 --ICHRDLKVDNLLLGADGRVCVGDFG 247
I HRDLK N+L+ +G + + DFG
Sbjct 151 KRIIHRDLKAGNVLMTLEGDIRLADFG 177
> CE24761
Length=305
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/144 (22%), Positives = 58/144 (40%), Gaps = 44/144 (30%)
Query 105 EDVKEEARILALLDHPRCLKLKDVL-DSNPESADGKVYFITNFLPGGALMVLNPMRDEEL 163
++ E I + L+HP +KL D+ K+Y + + PGG E+
Sbjct 73 HQLEREIEIQSHLNHPNIIKLYTYFWDAK------KIYLVLEYAPGG-----------EM 115
Query 164 VRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLGI 223
+ +S EP A + + +A + C ++ +
Sbjct 116 YKQLTVSKRFSEPTAA-----------------KYMYEIADALSYCHRKN---------V 149
Query 224 CHRDLKVDNLLLGADGRVCVGDFG 247
HRD+K +NLL+G+ G + +GDFG
Sbjct 150 IHRDIKPENLLIGSQGELKIGDFG 173
> At1g48260
Length=432
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 35/141 (24%), Positives = 57/141 (40%), Gaps = 42/141 (29%)
Query 107 VKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELVRS 166
+K E R L +L HP ++L +VL S K+Y + + GG L
Sbjct 56 IKREIRTLKVLKHPNIVRLHEVLASKT-----KIYMVLECVTGGDLF------------- 97
Query 167 CQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLGICHR 226
+ +V+ ++E R+ F + D + G+ HR
Sbjct 98 --------DRIVSKGKLSETQGRKMFQQLI----------------DGVSYCHNKGVFHR 133
Query 227 DLKVDNLLLGADGRVCVGDFG 247
DLK++N+LL A G + + DFG
Sbjct 134 DLKLENVLLDAKGHIKITDFG 154
> At3g08720
Length=471
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 54/146 (36%), Gaps = 48/146 (32%)
Query 105 EDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELV 164
E +K E IL +DHP ++LK + ++Y + +F+ GG L
Sbjct 183 EYMKAERDILTKIDHPFIVQLKYSFQTK-----YRLYLVLDFINGGHLFF---------- 227
Query 165 RSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL--- 221
QL F E A+ + + HL
Sbjct 228 ------------------------------QLYHQGLFREDLARVYTAEIVSAVSHLHEK 257
Query 222 GICHRDLKVDNLLLGADGRVCVGDFG 247
GI HRDLK +N+L+ DG V + DFG
Sbjct 258 GIMHRDLKPENILMDVDGHVMLTDFG 283
> At3g08730
Length=465
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 54/146 (36%), Gaps = 48/146 (32%)
Query 105 EDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEELV 164
E +K E IL +DHP ++LK + ++Y + +F+ GG L
Sbjct 177 EYMKAERDILTKIDHPFIVQLKYSFQTKY-----RLYLVLDFINGGHLFF---------- 221
Query 165 RSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL--- 221
QL F E A+ + + HL
Sbjct 222 ------------------------------QLYHQGLFREDLARVYTAEIVSAVSHLHEK 251
Query 222 GICHRDLKVDNLLLGADGRVCVGDFG 247
GI HRDLK +N+L+ DG V + DFG
Sbjct 252 GIMHRDLKPENILMDTDGHVMLTDFG 277
> SPBC1604.14c
Length=658
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 29/53 (54%), Gaps = 3/53 (5%)
Query 198 TLRTFTEAQAKCFIRDAAEVHEHL---GICHRDLKVDNLLLGADGRVCVGDFG 247
T T +E Q ++ E +HL GI HRD+K DN+LL G + + DFG
Sbjct 473 TNNTLSEGQIAAICKETLEGLQHLHENGIVHRDIKSDNILLSLQGDIKLTDFG 525
> At5g04510
Length=488
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 3/49 (6%)
Query 202 FTEAQAKCF---IRDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
+E +A+ + + DA E +G+ HRD+K +NLLL +DG + + DFG
Sbjct 136 LSEDEARFYTAEVVDALEYIHSMGLIHRDIKPENLLLTSDGHIKIADFG 184
> At4g30960
Length=441
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 59/144 (40%), Gaps = 43/144 (29%)
Query 104 MEDVKEEARILALLDHPRCLKLKDVLDSNPESADGKVYFITNFLPGGALMVLNPMRDEEL 163
++ +K E ++ ++ HP ++L +V+ S K+YF + GG L
Sbjct 66 VDQIKREISVMRMVKHPNIVELHEVMASK-----SKIYFAMELVRGGELFA--------- 111
Query 164 VRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETLRTFTEAQAKCFIRDAAEVHEHLGI 223
VA + ED+ R F QL + F C R G+
Sbjct 112 -------------KVAKGRLREDVARVYFQ-QLISAVDF------CHSR---------GV 142
Query 224 CHRDLKVDNLLLGADGRVCVGDFG 247
HRDLK +NLLL +G + V DFG
Sbjct 143 YHRDLKPENLLLDEEGNLKVTDFG 166
> ECU01g1320
Length=865
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 48/108 (44%), Gaps = 37/108 (34%)
Query 140 VYFITNFLPGGALMVLNPMRDEELVRSCQLSSDAPEPVVADMSVTEDIIREKFTPQLETL 199
+Y++ +F+PGG M L LS ED++ E +
Sbjct 116 LYYLMDFIPGGDFMGL-------------LSK-------------EDVLEEDWV------ 143
Query 200 RTFTEAQAKCFIRDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
F A+ + E+H+ LG HRDLK DN+L+G DG V + DFG
Sbjct 144 -RFYAAE---IVAALDELHK-LGWIHRDLKPDNVLIGIDGHVKLADFG 186
> 7300149
Length=569
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/26 (69%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 222 GICHRDLKVDNLLLGADGRVCVGDFG 247
GI HRD+K DN+LLG DG V V DFG
Sbjct 409 GIIHRDIKSDNVLLGMDGSVKVTDFG 434
> At3g10540
Length=483
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 3/49 (6%)
Query 202 FTEAQAKCF---IRDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
+E +A+ + + DA E ++G+ HRD+K +NLLL DG + + DFG
Sbjct 137 LSEDEARFYSAEVVDALEYIHNMGLIHRDIKPENLLLTLDGHIKIADFG 185
> Hs4759052
Length=809
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 190 EKFTPQLETLRTFTEAQAKCFIRDAAEVHEHL---GICHRDLKVDNLLLGADGRVCVGDF 246
E FT L FTE + + ++ + EHL GI +RD+K++N+LL ++G V + DF
Sbjct 138 ELFT-HLSQRERFTEHEVQIYVGEIVLALEHLHKLGIIYRDIKLENILLDSNGHVVLTDF 196
Query 247 G 247
G
Sbjct 197 G 197
Score = 32.3 bits (72), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 6/54 (11%)
Query 200 RTFTEAQAKCFIRD--AAEVHEH-LGICHRDLKVDNLLLGADG---RVCVGDFG 247
+ F+E +A +R +A H H LG+ HRDLK +NLL + + + DFG
Sbjct 514 KHFSETEASYIMRKLVSALSHMHDLGVVHRDLKPENLLFTDENDNLEIKIIDFG 567
> At4g22940
Length=458
Score = 42.4 bits (98), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 202 FTEAQAKCFIRDAAEVHEHLG---ICHRDLKVDNLLLGADGRVCVGDFG 247
F+E Q KC++R +H + HRD+K NLL+ DG + + DFG
Sbjct 200 FSEPQVKCYMRQLLRGLDHCHTNHVLHRDMKSSNLLINGDGVLKIADFG 248
> 7296346
Length=952
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 36/70 (51%), Gaps = 9/70 (12%)
Query 186 DIIREKFTPQLETLR----TFTEAQAKCF----IRDAAEVHEHLGICHRDLKVDNLLLGA 237
D + +ET++ +F + KC +R A +H++ I HRDLK NLLL
Sbjct 637 DYVEHDLKSLMETMKNRKQSFFPGEVKCLTQQLLRAVAHLHDNW-ILHRDLKTSNLLLSH 695
Query 238 DGRVCVGDFG 247
G + VGDFG
Sbjct 696 KGILKVGDFG 705
> SPAC1B9.02c
Length=696
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 30/49 (61%), Gaps = 3/49 (6%)
Query 202 FTEAQAKCFIRDAAEVHEHL---GICHRDLKVDNLLLGADGRVCVGDFG 247
F E +AK +I + EHL I +RDLK +N+LL ADG + + DFG
Sbjct 400 FPEQRAKFYIAELVLALEHLHKHDIIYRDLKPENILLDADGHIALCDFG 448
> 7294770
Length=1207
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 31/50 (62%), Gaps = 3/50 (6%)
Query 201 TFTEAQAKCFIRD---AAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
F E++ + +I + A E LGI +RD+K++N+LL +G + + DFG
Sbjct 360 NFEESRVRVYIAEVVLALEQLHQLGIIYRDIKLENILLDGEGHIVLSDFG 409
> 7299322
Length=434
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Query 200 RTFTEAQAKCFIRDAAEV----HEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
+ TE +A+ R A H+H ICHRDLK++N+LL G + DFG
Sbjct 175 KVLTEEEARRIFRQVATAVYYCHKH-KICHRDLKLENILLDEKGNAKIADFG 225
> Hs4502731
Length=360
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 32/57 (56%), Gaps = 6/57 (10%)
Query 196 LETLRT-FTEAQAKCFI----RDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDFG 247
LE + T F+EAQ KC + R +H + I HRDLKV NLL+ G V DFG
Sbjct 128 LENMPTPFSEAQVKCIVLQVLRGLQYLHRNF-IIHRDLKVSNLLMTDKGCVKTADFG 183
> CE18646
Length=528
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 36/61 (59%), Gaps = 4/61 (6%)
Query 190 EKFTPQLETLRTFTEAQAKCF---IRDAAEVHEHLGICHRDLKVDNLLLGADGRVCVGDF 246
E FT L+ +TF+EA+ + + I A H I +RD+K++NLLL DG + + DF
Sbjct 264 ELFT-HLQRCKTFSEARTRFYGSEIILALGYLHHRNIVYRDMKLENLLLDRDGHIKITDF 322
Query 247 G 247
G
Sbjct 323 G 323
> At5g44290
Length=644
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 29/49 (59%), Gaps = 3/49 (6%)
Query 202 FTEAQAKCFIRDAAEVHEH---LGICHRDLKVDNLLLGADGRVCVGDFG 247
F+E Q KC+++ H G+ HRD+K NLL+ ++G + + DFG
Sbjct 233 FSEPQVKCYMQQLLSGLHHCHSRGVLHRDIKGSNLLIDSNGVLKIADFG 281
Lambda K H
0.321 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5009866206
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40