bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3591_orf2
Length=291
Score E
Sequences producing significant alignments: (Bits) Value
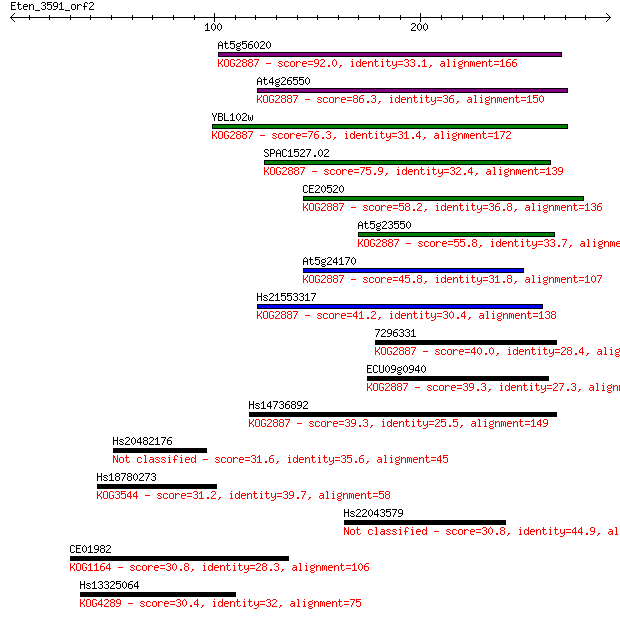
At5g56020 92.0 1e-18
At4g26550 86.3 7e-17
YBL102w 76.3 8e-14
SPAC1527.02 75.9 1e-13
CE20520 58.2 2e-08
At5g23550 55.8 1e-07
At5g24170 45.8 9e-05
Hs21553317 41.2 0.003
7296331 40.0 0.006
ECU09g0940 39.3 0.009
Hs14736892 39.3 0.010
Hs20482176 31.6 2.0
Hs18780273 31.2 2.8
Hs22043579 30.8 3.6
CE01982 30.8 3.8
Hs13325064 30.4 4.5
> At5g56020
Length=230
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 55/172 (31%), Positives = 96/172 (55%), Gaps = 7/172 (4%)
Query 102 QEQNGLLQSGLSAVKDGASRMLDGANSVV-----NATRATISDNPLSSQNMLMFGVVAAV 156
+E G L + + A+ + G +VV + ++ S P S + ++ FG++ A
Sbjct 46 EEDTGTLGFDIESAVRSANDTVSGTFNVVSKGVRDNLQSATSSMP-SGKALMYFGLLLAS 104
Query 157 GVLFMFLAF-LTLPLLVFAPSKFALLFTMGSLCFLAALALLRGVAALSRHLVEPSRLPFT 215
GV F+F+AF + LP++V P KFA+ FT+G + + LRG H+ RLP T
Sbjct 105 GVFFIFIAFTMFLPVMVLMPQKFAICFTLGCGFIIGSFFALRGPQNQLAHMSSAERLPLT 164
Query 216 AVYFSSLFLTLYATLMAKSYVLTLVFSVLQLCGLTSFLVSYIPGGSHMLKFM 267
+ +++ T+Y +++ SY+L+++FS LQ+ L + +SY PGGS +KF+
Sbjct 165 LGFIATMVGTIYVSMVLHSYILSVIFSALQVIALAYYCISYFPGGSSGMKFL 216
> At4g26550
Length=385
Score = 86.3 bits (212), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 54/167 (32%), Positives = 94/167 (56%), Gaps = 18/167 (10%)
Query 121 RMLDGANSVVNATRATISDNPLSSQNMLMFGVVAAVGVLFMFLAF-LTLPLLVFAPSKFA 179
R+ G + + ++ S P S + ++ FG++ A GV F+F+AF + LP++V P KFA
Sbjct 209 RVSKGVRDIPGSLQSATSSMP-SGKALMYFGLLLASGVFFIFIAFTMFLPVMVLMPQKFA 267
Query 180 LLFTMGSLCFLAALALLRG----VAALSR------------HLVEPSRLPFTAVYFSSLF 223
+ FT+G + + LRG +A +S H + P RLP T + +++
Sbjct 268 ICFTLGCGFIIGSFFALRGPKNQLAHMSSMEVCYTVLLCVCHALNPRRLPSTLGFIATMV 327
Query 224 LTLYATLMAKSYVLTLVFSVLQLCGLTSFLVSYIPGGSHMLKFMGQA 270
T+Y +++ SY+L+++FSVLQ+ L + +SY PGGS ++F+ A
Sbjct 328 GTIYVSMVLHSYILSVLFSVLQVLALVYYCISYFPGGSSGMRFLSSA 374
> YBL102w
Length=215
Score = 76.3 bits (186), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 54/183 (29%), Positives = 91/183 (49%), Gaps = 12/183 (6%)
Query 99 SRQQEQNGLLQSGLSAVKDGA----SRMLDGANSVVNATRATISDNPLS------SQNML 148
+RQQ G +S + A +R D ++ + + + D S ++ M+
Sbjct 23 TRQQNSQGFNESAKTLFSSWADSLNTRAQDIYQTLPVSRQDLVQDQEPSWFQLSRTERMV 82
Query 149 MF-GVVAAVGVLFMFLAFLTLPLLVFAPSKFALLFTMGSLCFLAALALLRGVAALSRHLV 207
+F + F FL P+L P KF LL+TMGSL F+ A +L G A +HL
Sbjct 83 LFVCFLLGATACFTLCTFL-FPVLAAKPRKFGLLWTMGSLLFVLAFGVLMGPLAYLKHLT 141
Query 208 EPSRLPFTAVYFSSLFLTLYATLMAKSYVLTLVFSVLQLCGLTSFLVSYIPGGSHMLKFM 267
RLPF+ +F++ F+T+Y +K+ VLT+ ++L+L + + +SY P G+ L+ +
Sbjct 142 ARERLPFSMFFFATCFMTIYFAAFSKNTVLTITCALLELVAVIYYAISYFPFGATGLRML 201
Query 268 GQA 270
A
Sbjct 202 SSA 204
> SPAC1527.02
Length=201
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 74/139 (53%), Gaps = 4/139 (2%)
Query 124 DGANSVVNATRATISDNPLSSQNMLMFGVVAAVGVLFMFLAFLTLPLLVFAPSKFALLFT 183
G NS ++ ++S + ++FG+ + +A P+LV P KF LL+T
Sbjct 49 SGGNSYFQSSEFSLSR----WERYMLFGICLLGSLACYAIACFMFPVLVLKPRKFVLLWT 104
Query 184 MGSLCFLAALALLRGVAALSRHLVEPSRLPFTAVYFSSLFLTLYATLMAKSYVLTLVFSV 243
MGSL + A+++G A R L RLP T YF +L T+ AT+ KS +L++VF V
Sbjct 105 MGSLLAVLGFAIVQGFVAHFRQLTTMERLPITLSYFVTLLATIIATIKIKSTILSIVFGV 164
Query 244 LQLCGLTSFLVSYIPGGSH 262
L + ++L+++ P G+
Sbjct 165 LHILSFVAYLIAFFPFGTR 183
> CE20520
Length=235
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 50/147 (34%), Positives = 79/147 (53%), Gaps = 11/147 (7%)
Query 143 SSQNMLMFGV----------VAAVGVLFMF-LAFLTLPLLVFAPSKFALLFTMGSLCFLA 191
S+Q+ MFG+ + +G +F F A + +P+++ + KFA L T+GSL L
Sbjct 81 STQDESMFGMTRTQRIIAFFMCIIGAIFCFSTAAVLIPVILVSTRKFAGLNTLGSLLLLL 140
Query 192 ALALLRGVAALSRHLVEPSRLPFTAVYFSSLFLTLYATLMAKSYVLTLVFSVLQLCGLTS 251
+ A L G + H+ P R T Y S+LF TLY++L KS + TL+ ++ Q L
Sbjct 141 SFAFLLGPKSYLTHMASPQRRLVTVSYLSALFATLYSSLWLKSTIFTLIAAIFQGFTLVW 200
Query 252 FLVSYIPGGSHMLKFMGQAAWQLLRKA 278
+++SY+PGG L FM LR++
Sbjct 201 YVLSYVPGGERGLFFMTSLFTSFLRRS 227
> At5g23550
Length=175
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 56/95 (58%), Gaps = 0/95 (0%)
Query 170 LLVFAPSKFALLFTMGSLCFLAALALLRGVAALSRHLVEPSRLPFTAVYFSSLFLTLYAT 229
L+ F P KF + FT+G+L L + A L G +++P+R+ TA+Y +S+ + L+
Sbjct 69 LVFFNPVKFGITFTLGNLMALGSTAFLIGPQRQVTMMLDPARIYATALYLASIIIALFCA 128
Query 230 LMAKSYVLTLVFSVLQLCGLTSFLVSYIPGGSHML 264
L ++ +LTL+ +L+ GL + +SYIP M+
Sbjct 129 LYVRNKLLTLLAIILEFTGLIWYSLSYIPFARTMV 163
> At5g24170
Length=138
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 57/108 (52%), Gaps = 6/108 (5%)
Query 143 SSQNMLMFGVVAAVGVLFMFLAFLTLPLLVF-APSKFALLFTMGSLCFLAALALLRGVAA 201
++Q M F A G+L MFL+ ++VF P KFALLFT G++ + + A L G
Sbjct 34 TTQRMYGFAASLATGLLLMFLS-----MIVFGIPIKFALLFTFGNVLAIGSTAFLMGPEQ 88
Query 202 LSRHLVEPSRLPFTAVYFSSLFLTLYATLMAKSYVLTLVFSVLQLCGL 249
+ +P R T++Y + + L L+ S +LT++ + ++C L
Sbjct 89 QMSMMFDPVRFLATSIYIGCVVVALICALLIHSKILTVLAILCEICAL 136
> Hs21553317
Length=159
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 42/142 (29%), Positives = 61/142 (42%), Gaps = 7/142 (4%)
Query 121 RMLDGANSVVNATRATISDNPLSSQN--MLMFGVVAAVGVLFMFLA--FLTLPLLVFAPS 176
R+L G + A + D S N + F + GV F L L LP
Sbjct 6 RVLSGQDDEEQGLTAQVLDASSLSFNTRLKWFAICFVCGVFFSILGTGLLWLP---GGIK 62
Query 177 KFALLFTMGSLCFLAALALLRGVAALSRHLVEPSRLPFTAVYFSSLFLTLYATLMAKSYV 236
FA+ +T+G+L LA+ L G + + E +RL T V TL A L
Sbjct 63 LFAVFYTLGNLAALASTCFLMGPVKQLKKMFEATRLLATIVMLLCFIFTLCAALWWHKKG 122
Query 237 LTLVFSVLQLCGLTSFLVSYIP 258
L ++F +LQ +T + +SYIP
Sbjct 123 LAVLFCILQFLSMTWYSLSYIP 144
> 7296331
Length=163
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 46/88 (52%), Gaps = 0/88 (0%)
Query 178 FALLFTMGSLCFLAALALLRGVAALSRHLVEPSRLPFTAVYFSSLFLTLYATLMAKSYVL 237
FA+ +T+G++ +A+ L G + + +RL T + + LT A ++ K L
Sbjct 67 FAVFYTLGNVISMASTCFLMGPFKQIKKMFAETRLIATIIVLVMMVLTFIAAIVWKKAGL 126
Query 238 TLVFSVLQLCGLTSFLVSYIPGGSHMLK 265
TL+F ++Q +T + +SYIP +K
Sbjct 127 TLIFIIIQSLAMTWYSLSYIPYARDAVK 154
> ECU09g0940
Length=171
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 40/88 (45%), Gaps = 0/88 (0%)
Query 174 APSKFALLFTMGSLCFLAALALLRGVAALSRHLVEPSRLPFTAVYFSSLFLTLYATLMAK 233
+PS F L +T+ + F L G + L + ++ + LTLY L
Sbjct 71 SPSGFILPYTISNFLFFIMFGFLLGFRSYLEGLFSKKKRVHSSWFIGCTLLTLYVVLKYD 130
Query 234 SYVLTLVFSVLQLCGLTSFLVSYIPGGS 261
Y+L L F +Q+ F +++IPGG+
Sbjct 131 RYLLNLAFCFIQVVSFIMFSLTFIPGGT 158
> Hs14736892
Length=160
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 38/155 (24%), Positives = 72/155 (46%), Gaps = 10/155 (6%)
Query 117 DGASRMLDGANSVVNATRATISDNPLSSQNMLMFGVVA--AVGVLFMFLAFLTLPLLVFA 174
D ++L G ++ + + + + S + + G +A A+G+L L +L++
Sbjct 2 DKLKKVLSGQDTEDRSGLSEVVEASSLSWSTRIKGFIACFAIGILCSLLG----TVLLWV 57
Query 175 PSK----FALLFTMGSLCFLAALALLRGVAALSRHLVEPSRLPFTAVYFSSLFLTLYATL 230
P K FA+ +T G++ + + L G + + EP+RL T + LTL +
Sbjct 58 PRKGLHLFAVFYTFGNIASIGSTIFLMGPVKQLKRMFEPTRLIATIMVLLCFALTLCSAF 117
Query 231 MAKSYVLTLVFSVLQLCGLTSFLVSYIPGGSHMLK 265
+ L L+F +LQ LT + +S+IP +K
Sbjct 118 WWHNKGLALIFCILQSLALTWYSLSFIPFARDAVK 152
> Hs20482176
Length=112
Score = 31.6 bits (70), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 51 TGGKPAVPGFLGNILGTKAGAAAPEGMGAGAGIWATPGRLPSGPM 95
G +P VP L ++L TK+G+ AP+ + W LPS P+
Sbjct 26 PGSRPCVPCSLPHLLRTKSGSPAPKSWNPFSASWGPRPPLPSRPV 70
> Hs18780273
Length=957
Score = 31.2 bits (69), Expect = 2.8, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 30/65 (46%), Gaps = 9/65 (13%)
Query 43 GTPMRGGSTGGKPAVPGFLGN--ILGTKAGAAAPEGMGAGAGIWATPGRL-----PSGPM 95
G P + G T G+P +PGF GN ++G K G P G G PG + P P
Sbjct 593 GAPGQDG-TRGEPGIPGFPGNRGLMGQK-GEIGPPGQQGKKGAPGMPGLMGSNGSPGQPG 650
Query 96 KPSSR 100
P S+
Sbjct 651 TPGSK 655
> Hs22043579
Length=215
Score = 30.8 bits (68), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 48/81 (59%), Gaps = 5/81 (6%)
Query 163 LAFLTLPLLVFAPSKFALLFTMGSLCFLAALALLRGVAALSRHL---VEPSRLPFTAVYF 219
LA L P+L+ KFALL+++GS LA ALLRG AA R L PSR +Y
Sbjct 93 LAALYAPVLLLRARKFALLWSLGSALALAGSALLRGGAACGRLLRCEEAPSRPAL--LYM 150
Query 220 SSLFLTLYATLMAKSYVLTLV 240
++L TL+A L +S +LT++
Sbjct 151 AALGATLFAALGLRSTLLTVL 171
> CE01982
Length=752
Score = 30.8 bits (68), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 42/113 (37%), Gaps = 7/113 (6%)
Query 30 QQEKKGYAMVSLDGTPMRGGSTGGKPAVPGFLGNILGTKAGAAAPEGMGA--GAGIWATP 87
Q KK A+ + G G KP P + N +K + M GA
Sbjct 17 QTSKKRKAVAASKARTPEGSGRGKKPTTPNGVANPTKSKTKVSTASTMNEKKGASTGDMK 76
Query 88 GRLPSGPMKPSSRQQEQNGLLQSGLSAVK-----DGASRMLDGANSVVNATRA 135
+ PS KPS + NG + G++ K +SR+LD VV A
Sbjct 77 KKEPSSRKKPSPMKCFANGEIVEGVNQFKVLSQIRKSSRLLDHDVYVVKDVEA 129
> Hs13325064
Length=2923
Score = 30.4 bits (67), Expect = 4.5, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 31/76 (40%), Gaps = 4/76 (5%)
Query 35 GYAMVSLDGTPMRGGSTGGKPAVPGFLGNILGTKAGAAAPEG-MGAGAGIWATPGRLPSG 93
G + L TP GG GK PG G +G APE + + G L G
Sbjct 2768 GAERLPLHSTPKDGGPGPGKAPWPGDFGTTAKESSGNGAPEERLRENGDALSREGSL--G 2825
Query 94 PMKPSSRQQEQNGLLQ 109
P+ P S Q G+L+
Sbjct 2826 PL-PGSSAQPHKGILK 2840
Lambda K H
0.322 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6460838058
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40