bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3585_orf2
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
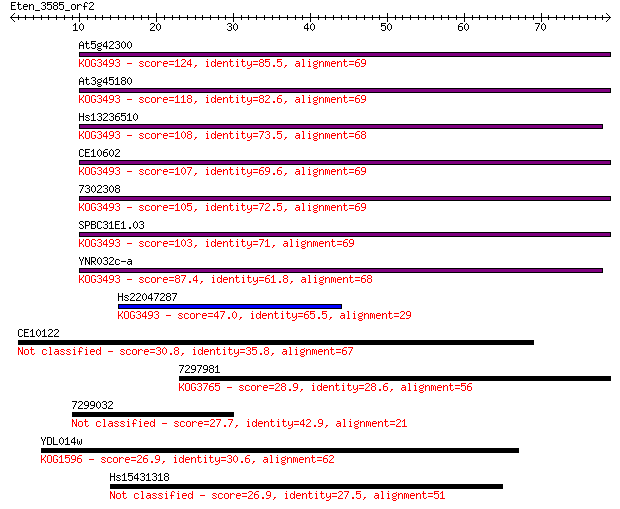
At5g42300 124 6e-29
At3g45180 118 2e-27
Hs13236510 108 2e-24
CE10602 107 7e-24
7302308 105 2e-23
SPBC31E1.03 103 8e-23
YNR032c-a 87.4 5e-18
Hs22047287 47.0 9e-06
CE10122 30.8 0.72
7297981 28.9 2.7
7299032 27.7 5.2
YDL014w 26.9 8.3
Hs15431318 26.9 10.0
> At5g42300
Length=73
Score = 124 bits (310), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 59/73 (80%), Positives = 66/73 (90%), Gaps = 4/73 (5%)
Query 10 MIEVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQ----CDKIRIQKWYTVYKDHITLEDY 65
MIEV+LNDRLG+K+RVKCN DDT+GDLKKLVAAQ +KIRIQKWY +YKDHITL+DY
Sbjct 1 MIEVVLNDRLGKKVRVKCNDDDTIGDLKKLVAAQTGTRAEKIRIQKWYNIYKDHITLKDY 60
Query 66 EIHDGMGLELYYN 78
EIHDGMGLELYYN
Sbjct 61 EIHDGMGLELYYN 73
> At3g45180
Length=73
Score = 118 bits (296), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 57/73 (78%), Positives = 65/73 (89%), Gaps = 4/73 (5%)
Query 10 MIEVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQC----DKIRIQKWYTVYKDHITLEDY 65
MIEV+LNDRLG+K+RVKCN +DT+GDLKKLVAAQ +KIRIQKWY +YKDHI L+DY
Sbjct 1 MIEVVLNDRLGKKVRVKCNEEDTIGDLKKLVAAQTGTRPEKIRIQKWYNIYKDHIPLKDY 60
Query 66 EIHDGMGLELYYN 78
EIHDGMGLELYYN
Sbjct 61 EIHDGMGLELYYN 73
> Hs13236510
Length=73
Score = 108 bits (271), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 50/72 (69%), Positives = 62/72 (86%), Gaps = 4/72 (5%)
Query 10 MIEVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQC----DKIRIQKWYTVYKDHITLEDY 65
MIEV+ NDRLG+K+RVKCN DDT+GDLKKL+AAQ +KI ++KWYT++KDH++L DY
Sbjct 1 MIEVVCNDRLGKKVRVKCNTDDTIGDLKKLIAAQTGTRWNKIVLKKWYTIFKDHVSLGDY 60
Query 66 EIHDGMGLELYY 77
EIHDGM LELYY
Sbjct 61 EIHDGMNLELYY 72
> CE10602
Length=73
Score = 107 bits (266), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 48/73 (65%), Positives = 60/73 (82%), Gaps = 4/73 (5%)
Query 10 MIEVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQC----DKIRIQKWYTVYKDHITLEDY 65
MIE+ +NDRLG+K+R+KCNP DT+GDLKKL+AAQ +KI ++KWYT+YKDHITL DY
Sbjct 1 MIEITVNDRLGKKVRIKCNPSDTIGDLKKLIAAQTGTRWEKIVLKKWYTIYKDHITLMDY 60
Query 66 EIHDGMGLELYYN 78
EIH+G ELYY
Sbjct 61 EIHEGFNFELYYQ 73
> 7302308
Length=73
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 50/73 (68%), Positives = 60/73 (82%), Gaps = 4/73 (5%)
Query 10 MIEVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQC----DKIRIQKWYTVYKDHITLEDY 65
MIE+ NDRLG+K+RVKCNPDDT+GDLKKL+AAQ +KI ++KWYT++KD I L DY
Sbjct 1 MIEITCNDRLGKKVRVKCNPDDTIGDLKKLIAAQTGTKHEKIVLKKWYTIFKDPIRLSDY 60
Query 66 EIHDGMGLELYYN 78
EIHDGM LELYY
Sbjct 61 EIHDGMNLELYYQ 73
> SPBC31E1.03
Length=73
Score = 103 bits (257), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 49/73 (67%), Positives = 61/73 (83%), Gaps = 4/73 (5%)
Query 10 MIEVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQCD----KIRIQKWYTVYKDHITLEDY 65
MIEV+ NDRLG+K+RVKC PDDTVGD KKLVAAQ +I ++KW++V+KD+ITL DY
Sbjct 1 MIEVLCNDRLGKKVRVKCMPDDTVGDFKKLVAAQTGTDPRRIVLKKWHSVFKDNITLADY 60
Query 66 EIHDGMGLELYYN 78
EIHDGM LE+YY+
Sbjct 61 EIHDGMSLEMYYS 73
> YNR032c-a
Length=73
Score = 87.4 bits (215), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 42/72 (58%), Positives = 56/72 (77%), Gaps = 4/72 (5%)
Query 10 MIEVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQC----DKIRIQKWYTVYKDHITLEDY 65
MIEV++NDRLG+K+RVKC +D+VGD KK+++ Q +KI +QK +V KDHI+LEDY
Sbjct 1 MIEVVVNDRLGKKVRVKCLAEDSVGDFKKVLSLQIGTQPNKIVLQKGGSVLKDHISLEDY 60
Query 66 EIHDGMGLELYY 77
E+HD LELYY
Sbjct 61 EVHDQTNLELYY 72
> Hs22047287
Length=279
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 19/29 (65%), Positives = 26/29 (89%), Gaps = 0/29 (0%)
Query 15 LNDRLGRKIRVKCNPDDTVGDLKKLVAAQ 43
L+D LG+K+R+KCN DDT+GD KKL+A+Q
Sbjct 219 LHDFLGKKVRIKCNTDDTIGDPKKLIASQ 247
> CE10122
Length=702
Score = 30.8 bits (68), Expect = 0.72, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 36/87 (41%), Gaps = 20/87 (22%)
Query 2 RFPPPEAKMIEVILND----RLGRKIRV----------------KCNPDDTVGDLKKLVA 41
RFPP + M +++ + LG IR KC G+L+KL
Sbjct 474 RFPPGKGNMSILMIGNSYVMNLGEHIRAHFNYNYSDYRYIAISGKCKTFGKFGNLRKLGL 533
Query 42 AQCDKIRIQKWYTVYKDHITLEDYEIH 68
+RI WY V D+ +L+ EI+
Sbjct 534 KISKFLRILLWYLVKLDYYSLKILEIY 560
> 7297981
Length=407
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 27/56 (48%), Gaps = 2/56 (3%)
Query 23 IRVKCNPDDTVGDLKKLVAAQCDKIRIQKWYTVYKDHITLEDYEIHDGMGLELYYN 78
+ + C + T+ LKK V C+ + +YKD+ LE+Y + D + Y N
Sbjct 18 LEISCLENGTLAKLKKFV--NCESKAYKSSNAIYKDYWVLENYVMADHGDIPCYSN 71
> 7299032
Length=523
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 9/21 (42%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 9 KMIEVILNDRLGRKIRVKCNP 29
K +V++ RLG K++ +CNP
Sbjct 23 KSTQVVVQSRLGEKMQTQCNP 43
> YDL014w
Length=327
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 35/66 (53%), Gaps = 6/66 (9%)
Query 5 PPEAKMI----EVILNDRLGRKIRVKCNPDDTVGDLKKLVAAQCDKIRIQKWYTVYKDHI 60
P +A++I + L D+ G I +K N D+ D + + A + K+R ++ + + +
Sbjct 247 PDQARIIALNSHMFLKDQGGVVISIKANCIDSTVDAETVFAREVQKLREERIKPL--EQL 304
Query 61 TLEDYE 66
TLE YE
Sbjct 305 TLEPYE 310
> Hs15431318
Length=456
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 14/59 (23%), Positives = 32/59 (54%), Gaps = 8/59 (13%)
Query 14 ILNDRLGRKIRVKCNPDDTV------GDLKKLVAAQCDKIR--IQKWYTVYKDHITLED 64
IL +LG K+R++ + + T+ G+++ A + R +++W+ + I+L+D
Sbjct 248 ILRSQLGEKLRIELDIEPTIDLNRVLGEMRAQYEAMLETNRQDVEQWFQAQSEGISLQD 306
Lambda K H
0.322 0.143 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171925608
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40