bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3584_orf1
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
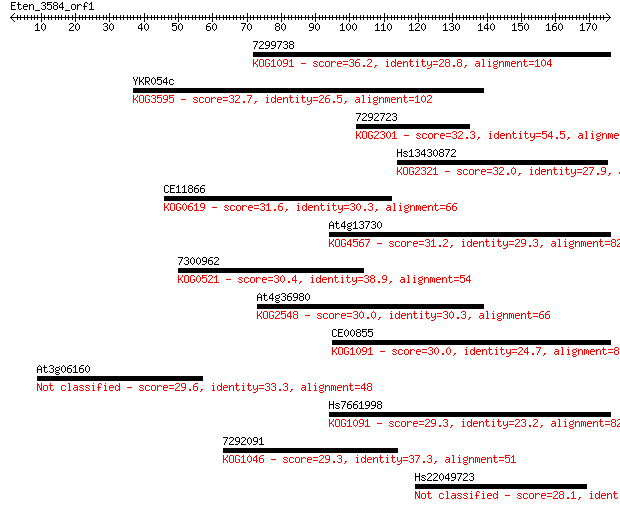
7299738 36.2 0.035
YKR054c 32.7 0.40
7292723 32.3 0.45
Hs13430872 32.0 0.61
CE11866 31.6 0.84
At4g13730 31.2 1.2
7300962 30.4 2.1
At4g36980 30.0 2.4
CE00855 30.0 2.8
At3g06160 29.6 3.2
Hs7661998 29.3 4.3
7292091 29.3 4.7
Hs22049723 28.1 9.8
> 7299738
Length=654
Score = 36.2 bits (82), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 51/104 (49%), Gaps = 13/104 (12%)
Query 72 EAAVAGDSSGTPAQGERLVVPFLRRLHWARLLGLLSGDTLEEWAEQVKEQRQQYESLRRQ 131
++A AGD L + R +HWA LL +L+ + W Q +QR +Y+ R
Sbjct 64 QSAFAGD----------LKMSKFRSVHWALLLRVLTSEH-RSWTSQRLQQRVRYDKFRAD 112
Query 132 QQLSASQLAALDPQRFHPLAATAGNPWSQKQADEELQEEIWRDI 175
+ QL A+D PL+ + + W+Q +D++L I +D+
Sbjct 113 YVRNPHQL-AVDCND-DPLSQSTQSVWNQYFSDQDLFAVIRQDV 154
> YKR054c
Length=4092
Score = 32.7 bits (73), Expect = 0.40, Method: Composition-based stats.
Identities = 27/113 (23%), Positives = 52/113 (46%), Gaps = 12/113 (10%)
Query 37 HSTANFPLKTLAEHV---LYGDAVDRQQLDIVRLQRQLEAAVAGDSSGTPAQGERLVVPF 93
+ST N P + +H+ +YG +D ++ D+ + + G + G R+ P
Sbjct 3975 NSTNNIPWAQVRDHIATIVYGGKIDEEK-DLEVVAKLCAHVFCGSDNLQIVPGVRIPQPL 4033
Query 94 LRRLH---WARLLGLLS-----GDTLEEWAEQVKEQRQQYESLRRQQQLSASQ 138
L++ ARL +LS D+L W + +E YE L+ ++ S+++
Sbjct 4034 LQQSEEEERARLTAILSNTIEPADSLSSWLQLPRESILNYERLQAKEVASSTE 4086
> 7292723
Length=1848
Score = 32.3 bits (72), Expect = 0.45, Method: Composition-based stats.
Identities = 18/33 (54%), Positives = 25/33 (75%), Gaps = 2/33 (6%)
Query 102 LLGLLSGDTLEEWAEQVKEQRQQYESLRRQQQL 134
+LG+LSG+ +E E+V E RQ++ LRRQQQL
Sbjct 308 VLGVLSGEFAKE-REKV-ENRQEFLKLRRQQQL 338
> Hs13430872
Length=638
Score = 32.0 bits (71), Expect = 0.61, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query 114 WAEQVKEQR---QQYESLRRQQQLSASQLAALDPQRFHPLAATAGNPWSQKQADEELQEE 170
W E+V++QR Q+ E ++RQ++L Q L PQ + A + ++L +
Sbjct 507 WVEEVRKQRRLLQREEKVKRQERLKEDQQTVLKPQFYEIKAGEEFRSFKDSATKQKLMNK 566
Query 171 IWRD 174
D
Sbjct 567 TLED 570
> CE11866
Length=961
Score = 31.6 bits (70), Expect = 0.84, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 46 TLAEHVLYGD---AVDRQQLDIVRLQ--RQLEAAVAGDSSGTPAQGERLVVPFLRRLHWA 100
T+ ++VL G +D+ LD + Q ++ A A + +G P +G+ V+P L +L
Sbjct 630 TMIQYVLNGGQIPGIDKATLDKIVKQTMNKMHTAAAANLAGNPVEGQEKVLPPLDKLPSG 689
Query 101 RLLGLLSGDTL 111
+ ++SG+ L
Sbjct 690 LVTQVMSGEPL 700
> At4g13730
Length=449
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 24/106 (22%), Positives = 46/106 (43%), Gaps = 25/106 (23%)
Query 94 LRRLHWARLLGLLSGDTLEEWAEQVKEQRQQYESLRRQQQLSASQLA----------ALD 143
+R + W LL LS D W+ ++ ++R QY+ + + ++ S++ + D
Sbjct 131 IRSIVWKLLLDYLSPDR-SLWSSELAKKRSQYKQFKEELLMNPSEVTRKMDKSKGGDSND 189
Query 144 PQ--------------RFHPLAATAGNPWSQKQADEELQEEIWRDI 175
P+ HPL+ + W+ D E+ E+I RD+
Sbjct 190 PKIESPGALSRSEITHEDHPLSLGTTSLWNNFFKDTEVLEQIERDV 235
> 7300962
Length=828
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 27/60 (45%), Gaps = 8/60 (13%)
Query 50 HVLYGDAVDRQQLDIVRLQRQLEAAVAGDSS------GTPAQGERLVVPFLRRLHWARLL 103
H+L D+ D L I LQ + AA+ DS+ T A L P RR+HW L
Sbjct 331 HILQADSADMLSLWISALQHSIGAAIQHDSTHHSRPQSTNAPNNSL--PAKRRIHWEEFL 388
> At4g36980
Length=550
Score = 30.0 bits (66), Expect = 2.4, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 37/68 (54%), Gaps = 2/68 (2%)
Query 73 AAVAGDSSGTPAQGERLVVP--FLRRLHWARLLGLLSGDTLEEWAEQVKEQRQQYESLRR 130
+ VAG SS +Q E+ P L+R+ +L + D+ E A++ +++RQ+ E L
Sbjct 361 SKVAGTSSSKQSQAEKKETPQERLKRIMNKQLTKQIKKDSATETAKKREQERQRLEKLAE 420
Query 131 QQQLSASQ 138
+LS ++
Sbjct 421 TSRLSRNR 428
> CE00855
Length=585
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 20/82 (24%), Positives = 37/82 (45%), Gaps = 2/82 (2%)
Query 95 RRLHWARLLGLLSGDTLEEWAEQVKEQRQQYESLRRQQQLSASQLA-ALDPQRFHPLAAT 153
R W +L L +T +W + R Y + + + + DP+ +PLA+
Sbjct 46 RSAVWRLVLRCLPYET-SDWEISLSRSRNLYRAHKENHLIDPHDTKFSQDPEFNNPLASI 104
Query 154 AGNPWSQKQADEELQEEIWRDI 175
NPW+ D +L++ I +D+
Sbjct 105 EQNPWNTFFEDNDLRDIIGKDV 126
> At3g06160
Length=330
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 9 MDDTAPEGEVGLLQLTPDVLPALSRAVEHSTANFPLKTLAEHVLYGDA 56
+ D + V L + L +LS HST+N L++L+ L+GDA
Sbjct 130 ISDDTEDDNVSLHSPSNVSLDSLSNDSHHSTSNVSLRSLSNDSLHGDA 177
> Hs7661998
Length=795
Score = 29.3 bits (64), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 19/83 (22%), Positives = 40/83 (48%), Gaps = 2/83 (2%)
Query 94 LRRLHWARLLGLLSGDTLEEWAEQVKEQRQQYESLRRQQQLSASQLAAL-DPQRFHPLAA 152
R + W L +L D +W +++E R Y +++ + ++ D +PL+
Sbjct 86 FRSICWKLFLCVLPQDK-SQWISRIEELRAWYSNIKEIHITNPRKVVGQQDLMINNPLSQ 144
Query 153 TAGNPWSQKQADEELQEEIWRDI 175
G+ W++ D+EL+ I +D+
Sbjct 145 DEGSLWNKFFQDKELRSMIEQDV 167
> 7292091
Length=866
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 22/51 (43%), Gaps = 0/51 (0%)
Query 63 DIVRLQRQLEAAVAGDSSGTPAQGERLVVPFLRRLHWARLLGLLSGDTLEE 113
D R R L VA P GE + LR L RL+ S DT+EE
Sbjct 628 DFHRFGRNLYEPVAYRLGWEPRDGENHLDTLLRSLVLTRLVSFRSSDTIEE 678
> Hs22049723
Length=280
Score = 28.1 bits (61), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 119 KEQRQQYESLRRQQQLSASQLAALDPQRFHPLAATAGNPWSQKQADEELQ 168
K + Q Y S+ + + + SQ+ +RFH + + W + EE+Q
Sbjct 214 KRRFQLYRSMNSRARKNRSQIVLFQKRRFHFFCSMSCRAWVSPEELEEIQ 263
Lambda K H
0.315 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2671071884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40