bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3567_orf3
Length=191
Score E
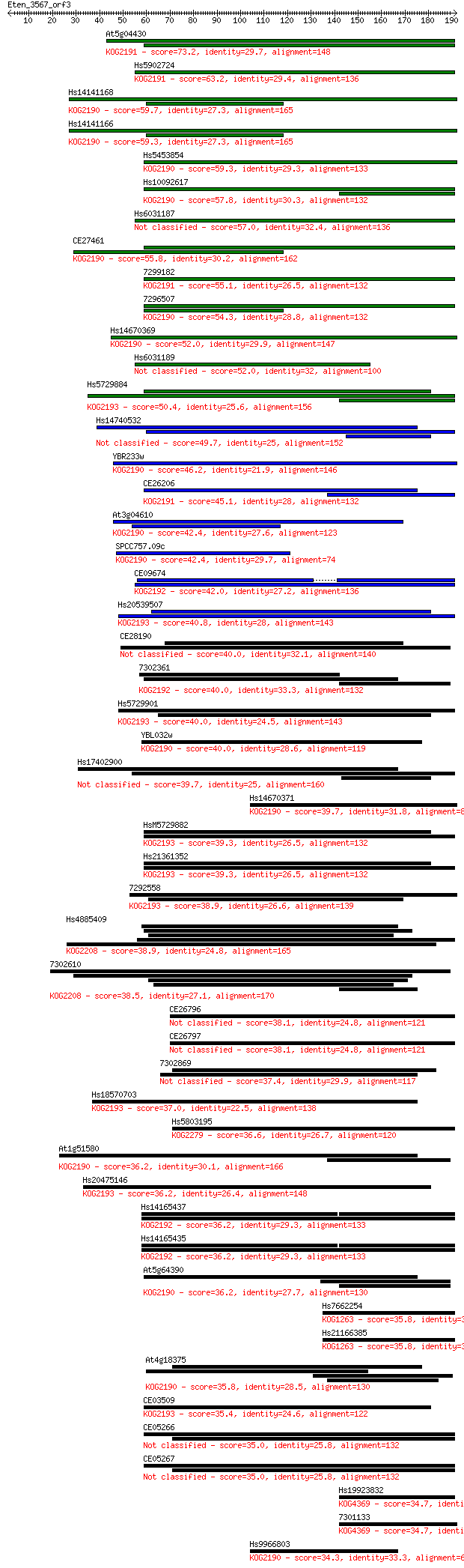
Sequences producing significant alignments: (Bits) Value
At5g04430 73.2 3e-13
Hs5902724 63.2 3e-10
Hs14141168 59.7 4e-09
Hs14141166 59.3 4e-09
Hs5453854 59.3 4e-09
Hs10092617 57.8 1e-08
Hs6031187 57.0 2e-08
CE27461 55.8 5e-08
7299182 55.1 8e-08
7296507 54.3 1e-07
Hs14670369 52.0 7e-07
Hs6031189 52.0 8e-07
Hs5729884 50.4 2e-06
Hs14740532 49.7 4e-06
YBR233w 46.2 4e-05
CE26206 45.1 8e-05
At3g04610 42.4 5e-04
SPCC757.09c 42.4 6e-04
CE09674 42.0 8e-04
Hs20539507 40.8 0.002
CE28190 40.0 0.003
7302361 40.0 0.003
Hs5729901 40.0 0.003
YBL032w 40.0 0.003
Hs17402900 39.7 0.004
Hs14670371 39.7 0.004
HsM5729882 39.3 0.005
Hs21361352 39.3 0.005
7292558 38.9 0.006
Hs4885409 38.9 0.006
7302610 38.5 0.009
CE26796 38.1 0.010
CE26797 38.1 0.011
7302869 37.4 0.018
Hs18570703 37.0 0.023
Hs5803195 36.6 0.028
At1g51580 36.2 0.037
Hs20475146 36.2 0.039
Hs14165437 36.2 0.039
Hs14165435 36.2 0.040
At5g64390 36.2 0.040
Hs7662254 35.8 0.055
Hs21166385 35.8 0.057
At4g18375 35.8 0.059
CE03509 35.4 0.069
CE05266 35.0 0.092
CE05267 35.0 0.097
Hs19923832 34.7 0.11
7301133 34.7 0.11
Hs9966803 34.3 0.14
> At5g04430
Length=313
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 78/155 (50%), Gaps = 7/155 (4%)
Query 43 DPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPES 102
+P + S P H++ L+ A A S+IGK +TIT + G + + ++ FP +
Sbjct 21 EPHDSSEADSAEKPTHIRFLVSNAAAGSVIGKGGSTITEFQAKSGARIQLSRNQEFFPGT 80
Query 103 TDRILLMSGSTESISTGIIALLERC-KEVHP---NLPYDQMYAKLVVPTSVCPTVIGPGG 158
TDRI+++SGS + + G+ +L++ E+H N + +LVVP S C +IG GG
Sbjct 81 TDRIIMISGSIKEVVNGLELILDKLHSELHAEDGNEVEPRRRIRLVVPNSSCGGIIGKGG 140
Query 159 TRVREIREATLTRITV---DRNSSPCPERIIAVHG 190
++ E + I + D +R++ + G
Sbjct 141 ATIKSFIEESKAGIKISPLDNTFYGLSDRLVTLSG 175
Score = 35.0 bits (79), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 40/166 (24%), Positives = 63/166 (37%), Gaps = 34/166 (20%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
++L++ + +IGK ATI EE + + N F +DR++ +SG+ E
Sbjct 123 IRLVVPNSSCGGIIGKGGATIKSFIEESKAGIKISPLDNTFYGLSDRLVTLSGTFEEQMR 182
Query 119 GIIALLERCKE-------VHPNLPYDQMY--------------------AKLVVPTSVCP 151
I +L + E VH Y Y A V V
Sbjct 183 AIDLILAKLTEDDHYSQNVHSPYSYAAGYNSVNYAPNGSGGKYQNHKEEASTTVTIGVAD 242
Query 152 T----VIGPGGTRVREIREATLTRITV-DRNS--SPCPERIIAVHG 190
V+G GG + EI + T RI + DR S +R +++ G
Sbjct 243 EHIGLVLGRGGRNIMEITQMTGARIKISDRGDFMSGTTDRKVSITG 288
> Hs5902724
Length=492
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/154 (25%), Positives = 75/154 (48%), Gaps = 18/154 (11%)
Query 55 GPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTE 114
G + +K+LI A S+IGK TI ++ +E G ++ + SK+ +P +T+R+ L+ G+ E
Sbjct 31 GEYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKDFYPGTTERVCLVQGTAE 90
Query 115 SISTGIIALLERCKEVHPNL----------------PYDQMYAKLVVPTSVCPTVIGPGG 158
+++ + E+ +E+ + P AKL+VP S +IG GG
Sbjct 91 ALNAVHSFIAEKVREIPQAMTKPEVVNILQPQTTMNPDRAKQAKLIVPNSTAGLIIGKGG 150
Query 159 TRVREIREATLTRITVDRNSSPC--PERIIAVHG 190
V+ + E + + + + ER++ V G
Sbjct 151 ATVKAVMEQSGAWVQLSQKPEGINLQERVVTVSG 184
> Hs14141168
Length=366
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 45/173 (26%), Positives = 86/173 (49%), Gaps = 31/173 (17%)
Query 27 ETGVLQEGMNMEALKEDPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEEC 86
+TGV++ G+N+ ++LL+ S+IGK+ ++ ++ EE
Sbjct 2 DTGVIEGGLNVT------------------LTIRLLMHGKEVGSIIGKKGESVKKMREES 43
Query 87 GCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKEVHPNLPYDQMYA----- 141
G +++ S+ PE RI+ ++G T +I ++++ +E + + A
Sbjct 44 GARINI--SEGNCPE---RIITLAGPTNAIFKAFAMIIDKLEEDISSSMTNSTAASRPPV 98
Query 142 --KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSP-CPERIIAVHGM 191
+LVVP S C ++IG GG +++EIRE+T ++ V + P ER I + G+
Sbjct 99 TLRLVVPASQCGSLIGKGGCKIKEIRESTGAQVQVAGDMLPNSTERAITIAGI 151
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 3/58 (5%)
Query 60 KLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
+L I L +IG++ A I + + G + + N STDR + ++GS SIS
Sbjct 292 ELTIPNDLIGCIIGRQGAKINEIRQMSGAQIKI---ANPVEGSTDRQVTITGSAASIS 346
> Hs14141166
Length=362
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 45/173 (26%), Positives = 86/173 (49%), Gaps = 31/173 (17%)
Query 27 ETGVLQEGMNMEALKEDPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEEC 86
+TGV++ G+N+ ++LL+ S+IGK+ ++ ++ EE
Sbjct 2 DTGVIEGGLNVT------------------LTIRLLMHGKEVGSIIGKKGESVKKMREES 43
Query 87 GCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKEVHPNLPYDQMYA----- 141
G +++ S+ PE RI+ ++G T +I ++++ +E + + A
Sbjct 44 GARINI--SEGNCPE---RIITLAGPTNAIFKAFAMIIDKLEEDISSSMTNSTAASRPPV 98
Query 142 --KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSP-CPERIIAVHGM 191
+LVVP S C ++IG GG +++EIRE+T ++ V + P ER I + G+
Sbjct 99 TLRLVVPASQCGSLIGKGGCKIKEIRESTGAQVQVAGDMLPNSTERAITIAGI 151
Score = 28.5 bits (62), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 3/58 (5%)
Query 60 KLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
+L I L +IG++ A I + + G + + N STDR + ++GS SIS
Sbjct 288 ELTIPNDLIGCIIGRQGAKINEIRQMSGAQIKI---ANPVEGSTDRQVTITGSAASIS 342
> Hs5453854
Length=356
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 78/141 (55%), Gaps = 13/141 (9%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
++LL+ S+IGK+ ++ R+ EE G +++ S+ PE RI+ ++G T +I
Sbjct 16 IRLLMHGKEVGSIIGKKGESVKRIREESGARINI--SEGNCPE---RIITLTGPTNAIFK 70
Query 119 GIIALLERCKE-VHPNLPYDQMYAK------LVVPTSVCPTVIGPGGTRVREIREATLTR 171
++++ +E ++ ++ ++ LVVP + C ++IG GG +++EIRE+T +
Sbjct 71 AFAMIIDKLEEDINSSMTNSTAASRPPVTLRLVVPATQCGSLIGKGGCKIKEIRESTGAQ 130
Query 172 ITVDRNSSP-CPERIIAVHGM 191
+ V + P ER I + G+
Sbjct 131 VQVAGDMLPNSTERAITIAGV 151
> Hs10092617
Length=339
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/140 (28%), Positives = 74/140 (52%), Gaps = 13/140 (9%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
++LL+ S+IGK+ T+ ++ EE G +++ S+ PE RI+ ++G T++I
Sbjct 16 IRLLMHGKEVGSIIGKKGETVKKMREESGARINI--SEGNCPE---RIVTITGPTDAIFK 70
Query 119 GIIALLERCKE-------VHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTR 171
+ + +E P + +LVVP S C ++IG GG++++EIRE+T +
Sbjct 71 AFAMIAYKFEEDIINSMSNSPATSKPPVTLRLVVPASQCGSLIGKGGSKIKEIRESTGAQ 130
Query 172 ITVDRNSSP-CPERIIAVHG 190
+ V + P ER + + G
Sbjct 131 VQVAGDMLPNSTERAVTISG 150
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHG 190
+L +P + +IG GT++ EIR+ + +I + + ER I + G
Sbjct 265 ELTIPNDLIGCIIGRQGTKINEIRQMSGAQIKIANATEGSSERQITITG 313
> Hs6031187
Length=486
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 80/159 (50%), Gaps = 25/159 (15%)
Query 55 GPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHV--LS-SKNKFPESTDRILLMSG 111
G + +K+LI A S+IGK TI ++ +E G ++ + LS SK+ +P +T+R+ L+ G
Sbjct 48 GQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKLSKSKDFYPGTTERVCLIQG 107
Query 112 STESIST--GIIALLERCKEVHPNL----------------PYDQMYAKLVVPTSVCPTV 153
+ E+++ G IA E+ +E+ N+ P K++VP S +
Sbjct 108 TVEALNAVHGFIA--EKIREMPQNVAKTEPVSILQPQTTVNPDRIKQVKIIVPNSTAGLI 165
Query 154 IGPGGTRVREIREATLTRITVDR--NSSPCPERIIAVHG 190
IG GG V+ + E + + + + + ER++ V G
Sbjct 166 IGKGGATVKAVMEQSGAWVQLSQKPDGINLQERVVTVSG 204
> CE27461
Length=451
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 72/137 (52%), Gaps = 10/137 (7%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
++LL+ S+IGK+ I ++ EE G +++ S PE RI+ ++G+ I
Sbjct 74 IRLLMQGKEVGSIIGKKGDQIKKIREESGAKINI--SDGSCPE---RIVTITGTLGVIGK 128
Query 119 GIIALLERCKE---VHPN-LPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITV 174
+ + +E + PN +P + +++VP + C ++IG GG+++++IREAT I V
Sbjct 129 AFNMVCNKFEEDMLLLPNSVPKPPITMRVIVPATQCGSLIGKGGSKIKDIREATGASIQV 188
Query 175 DRNSSP-CPERIIAVHG 190
P ER + + G
Sbjct 189 ASEMLPHSTERAVTLSG 205
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 46/91 (50%), Gaps = 5/91 (5%)
Query 29 GVLQEGMNM--EALKEDPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEEC 86
GV+ + NM +ED L V P P +++++ SLIGK + I + E
Sbjct 124 GVIGKAFNMVCNKFEEDMLLLPNSVPKP-PITMRVIVPATQCGSLIGKGGSKIKDIREAT 182
Query 87 GCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
G S+ V S P ST+R + +SG+ ++I+
Sbjct 183 GASIQVAS--EMLPHSTERAVTLSGTADAIN 211
> 7299182
Length=493
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 35/146 (23%), Positives = 75/146 (51%), Gaps = 15/146 (10%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
+K+L+ + ++IGK TI + ++ G + + S + +P +T+R+ L++GSTE+I
Sbjct 1 MKILVPAVASGAIIGKGGETIASLQKDTGARVKMSKSHDFYPGTTERVCLITGSTEAIMV 60
Query 119 GIIALLERCKEVHPNLPYD------------QMYAKLVVPTSVCPTVIGPGGTRVREIRE 166
+ ++++ +E P+L K++VP S +IG GG +++I+E
Sbjct 61 VMEFIMDKIRE-KPDLTNKIVDTDSKQTQERDKQVKILVPNSTAGMIIGKGGAFIKQIKE 119
Query 167 ATLTRITVDRNSSPCP--ERIIAVHG 190
+ + + + + + ER I + G
Sbjct 120 ESGSYVQISQKPTDVSLQERCITIIG 145
> 7296507
Length=386
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 38/139 (27%), Positives = 71/139 (51%), Gaps = 12/139 (8%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
++L++ S+IGK+ + R EE G +++ S PE RI+ +SG+T +I +
Sbjct 26 IRLIMQGKEVGSIIGKKGEIVNRFREESGAKINI--SDGSCPE---RIVTVSGTTNAIFS 80
Query 119 GIIALLER----CKEVHP--NLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRI 172
+ ++ C + + + Q+ +L+VP S C ++IG G++++EIR+ T I
Sbjct 81 AFTLITKKFEEWCSQFNDVGKVGKTQIPIRLIVPASQCGSLIGKSGSKIKEIRQTTGCSI 140
Query 173 TVDRNSSP-CPERIIAVHG 190
V P ER + + G
Sbjct 141 QVASEMLPNSTERAVTLSG 159
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
++L++ + SLIGK + I + + GCS+ V S P ST+R + +SGS E I+
Sbjct 109 IRLIVPASQCGSLIGKSGSKIKEIRQTTGCSIQVASEM--LPNSTERAVTLSGSAEQIT 165
> Hs14670369
Length=403
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 79/155 (50%), Gaps = 15/155 (9%)
Query 45 LEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTD 104
LE+E +S+ +++L+ S+IGK+ T+ R+ E+ S + S+ PE
Sbjct 8 LEEEPELSIT--LTLRMLMHGKEVGSIIGKKGETVKRIREQS--SARITISEGSCPE--- 60
Query 105 RILLMSGSTESISTGIIALLERCKE---VHP----NLPYDQMYAKLVVPTSVCPTVIGPG 157
RI ++GST ++ + + + E P N+ + +LV+P S C ++IG
Sbjct 61 RITTITGSTAAVFHAVSMIAFKLDEDLCAAPANGGNVSRPPVTLRLVIPASQCGSLIGKA 120
Query 158 GTRVREIREATLTRITVDRNSSP-CPERIIAVHGM 191
GT+++EIRE T ++ V + P ER + V G+
Sbjct 121 GTKIKEIRETTGAQVQVAGDLLPNSTERAVTVSGV 155
> Hs6031189
Length=181
Score = 52.0 bits (123), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 59/102 (57%), Gaps = 4/102 (3%)
Query 55 GPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTE 114
G + +K+LI A S+IGK TI ++ +E G ++ + SK+ +P +T+R+ L+ G+ E
Sbjct 48 GQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKDFYPGTTERVCLIQGTVE 107
Query 115 SIST--GIIALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVI 154
+++ G IA E+ +E+ N+ + + L T+V P I
Sbjct 108 ALNAVHGFIA--EKIREMPQNVAKTEPVSILQPQTTVNPDRI 147
> Hs5729884
Length=556
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/124 (26%), Positives = 64/124 (51%), Gaps = 4/124 (3%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
V L I ++IGK+ A I ++A G S+ + ++ P+ ++R+++++G E+
Sbjct 387 VNLFIPTQAVGAIIGKKGAHIKQLARFAGASIKIAPAEG--PDVSERMVIITGPPEAQFK 444
Query 119 GIIALLERCKEVHPNLPYDQ--MYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDR 176
+ + KE + P ++ + A + VP+S VIG GG V E++ T + V R
Sbjct 445 AQGRIFGKLKEENFFNPKEEVKLEAHIRVPSSTAGRVIGKGGKTVNELQNLTSAEVIVPR 504
Query 177 NSSP 180
+ +P
Sbjct 505 DQTP 508
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/185 (18%), Positives = 74/185 (40%), Gaps = 33/185 (17%)
Query 35 MNMEALKEDPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLS 94
M +E ++++ E + +P +K+L L LIGKE + ++ E G + + S
Sbjct 257 MILEIMQKEADETKLAEEIP----LKILAHNGLVGRLIGKEGRNLKKIEHETGTKITISS 312
Query 95 SKNKFPESTDRILLMSGSTESISTGIIALLERCKEVHPN--------------------- 133
++ + +R + + G+ E+ ++ I ++++ +E N
Sbjct 313 LQDLSIYNPERTITVKGTVEACASAEIEIMKKLREAFENDMLAVNTHSGYFSSLYPHHQF 372
Query 134 --LPYDQMY-----AKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSP-CPERI 185
P+ Y L +PT +IG G ++++ I + P ER+
Sbjct 373 GPFPHHHSYPEQEIVNLFIPTQAVGAIIGKKGAHIKQLARFAGASIKIAPAEGPDVSERM 432
Query 186 IAVHG 190
+ + G
Sbjct 433 VIITG 437
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRN-SSPCPERIIAVHG 190
+++VPT +IG G ++ I + T +R+ + R +S E+ + +H
Sbjct 197 RILVPTQFVGAIIGKEGLTIKNITKQTQSRVDIHRKENSGAAEKPVTIHA 246
> Hs14740532
Length=572
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/148 (22%), Positives = 71/148 (47%), Gaps = 15/148 (10%)
Query 39 ALKEDPLEDERPVSLPGPFHVKLLID------PALANSLIGKESATITRVAEECGCSMHV 92
+++ PL+D L H + +I + +IG+ I+R+ E GC + +
Sbjct 54 GVQKRPLDDGVGNQLGALVHQRTVITEEFKVPDKMVGFIIGRGGEQISRIQAESGCKIQI 113
Query 93 LSSKNKFPESTDRILLMSGSTESISTG---IIALLERCKE---VHPNLPYDQMYAKLVVP 146
S + PE R +++G+ ESI + +++RC+ H ++ + ++++P
Sbjct 114 ASESSGIPE---RPCVLTGTPESIEQAKRLLGQIVDRCRNGPGFHNDIDSNSTIQEILIP 170
Query 147 TSVCPTVIGPGGTRVREIREATLTRITV 174
S VIG GG +++++E T ++ +
Sbjct 171 ASKVGLVIGRGGETIKQLQERTGVKMVM 198
Score = 33.1 bits (74), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 32/141 (22%), Positives = 53/141 (37%), Gaps = 11/141 (7%)
Query 60 KLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTG 119
++LI + +IG+ TI ++ E G M V+ P D+ L ++G +
Sbjct 166 EILIPASKVGLVIGRGGETIKQLQERTGVKM-VMIQDGPLPTGADKPLRITGDAFKVQQA 224
Query 120 IIALLERCKEVHP----------NLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATL 169
+LE +E N ++ VP VIG G +++I+
Sbjct 225 REMVLEIIREKDQADFRGVRGDFNSRMGGGSIEVSVPRFAVGIVIGRNGEMIKKIQNDAG 284
Query 170 TRITVDRNSSPCPERIIAVHG 190
RI + PER V G
Sbjct 285 VRIQFKPDDGISPERAAQVMG 305
Score = 28.9 bits (63), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 145 VPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSP 180
VP C VIG GG ++ I + + + + RN P
Sbjct 361 VPADKCGLVIGKGGENIKSINQQSGAHVELQRNPPP 396
> YBR233w
Length=413
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/150 (21%), Positives = 75/150 (50%), Gaps = 7/150 (4%)
Query 46 EDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDR 105
+ + P ++P H+++L A+ ++G + ATI+R+ E +++ ++ PE R
Sbjct 56 DHDSPDNVPSDVHLRMLCLVKHASLIVGHKGATISRIKSETSARINISNNIRGVPE---R 112
Query 106 ILLMSGSTESISTGIIALLERCKEVHPNLPYD---QMYAKLVVPTSVCPTVIGPGGTRVR 162
I+ + G+ + ++ ++ E H N ++ L++P + +IG G+R+R
Sbjct 113 IVYVRGTCDDVAKAYGMIVRALLEEHGNEDNGEDIEISINLLIPHHLMGCIIGKRGSRLR 172
Query 163 EIREATLTRITVDRNSSPCP-ERIIAVHGM 191
EI + + ++ N +RI+ ++G+
Sbjct 173 EIEDLSAAKLFASPNQLLLSNDRILTINGV 202
> CE26206
Length=413
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 29/126 (23%), Positives = 58/126 (46%), Gaps = 10/126 (7%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
+K+LI ++IGK + + + C + + + +P +++RI L+ G +I
Sbjct 44 IKILIPSNAVGAIIGKGGEAMRNLKNDNNCRVQMSKNSETYPGTSERICLVKGRLNNIMA 103
Query 119 GIIALL----ERCKEVHPNLPYDQMYA------KLVVPTSVCPTVIGPGGTRVREIREAT 168
I ++ E+C + + +D K+V+P + VIG G +++IRE
Sbjct 104 VIESIQDKIREKCADQGGSDAFDHKNTSRGAEIKIVMPNTSAGMVIGKSGANIKDIREQF 163
Query 169 LTRITV 174
+I V
Sbjct 164 GCQIQV 169
Score = 36.6 bits (83), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 30/57 (52%), Gaps = 3/57 (5%)
Query 137 DQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCP---ERIIAVHG 190
D + K+++P++ +IG GG +R ++ R+ + +NS P ERI V G
Sbjct 40 DHLSIKILIPSNAVGAIIGKGGEAMRNLKNDNNCRVQMSKNSETYPGTSERICLVKG 96
> At3g04610
Length=577
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 34/134 (25%), Positives = 64/134 (47%), Gaps = 14/134 (10%)
Query 46 EDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDR 105
E++R PG ++L+ S+IG++ I ++ EE + +L P +T+R
Sbjct 176 EEKRWPGWPGETVFRMLVPAQKVGSIIGRKGDVIKKIVEETRARIKILDGP---PGTTER 232
Query 106 ILLMSGSTESIST------GIIALLERC-----KEVHPNLPYDQMYAKLVVPTSVCPTVI 154
+++SG E S+ G++ + R E P ++ +L+VP S ++I
Sbjct 233 AVMVSGKEEPESSLPPSMDGLLRVHMRIVDGLDGEASQAPPPSKVSTRLLVPASQAGSLI 292
Query 155 GPGGTRVREIREAT 168
G G V+ I+EA+
Sbjct 293 GKQGGTVKAIQEAS 306
Score = 36.6 bits (83), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 54 PGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKN--KFPESTDRILLMSG 111
P +LL+ + A SLIGK+ T+ + E C + VL S++ F DR++ + G
Sbjct 274 PSKVSTRLLVPASQAGSLIGKQGGTVKAIQEASACIVRVLGSEDLPVFALQDDRVVEVVG 333
Query 112 STESI 116
S+
Sbjct 334 EPTSV 338
> SPCC757.09c
Length=398
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query 47 DERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRI 106
D P P ++LLI +L S+IG+ I + ++C C M ++SK+ P+ST+R
Sbjct 169 DGTPSDANTPRKLRLLIAHSLMGSIIGRNGLRIKLIQDKCSCRM--IASKDMLPQSTERT 226
Query 107 LLMSGSTESISTGI 120
+ + G+ +++ I
Sbjct 227 VEIHGTVDNLHAAI 240
> CE09674
Length=397
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 42/75 (56%), Gaps = 2/75 (2%)
Query 56 PFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTES 115
P V++L+ + A +LIG+ + I + E+C + + + P STDR+L+ SG ++
Sbjct 124 PCEVRMLVHQSHAGALIGRNGSKIKELREKCSARLKIFTG--CAPGSTDRVLITSGEQKN 181
Query 116 ISTGIIALLERCKEV 130
+ I +++ KE+
Sbjct 182 VLGIIEEVMKELKEI 196
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 34/50 (68%), Gaps = 1/50 (2%)
Query 141 AKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHG 190
A++ +P+ + T+IG GG R+ IR+ + +IT+++ S+ PERII + G
Sbjct 319 AQVTIPSDLGGTIIGRGGERIARIRQESGAQITLEQ-SNGQPERIITIKG 367
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 64/142 (45%), Gaps = 18/142 (12%)
Query 55 GPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTE 114
G F V+LL+ A ++IGK I R+ E + HV + PE R+ ++ +
Sbjct 48 GKFEVRLLVSSKSAGAIIGKGGENIKRLRAE--FNAHVQVPDSNTPE---RVCTVTADEK 102
Query 115 SISTGIIALLERCKEVHPNL-----PYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATL 169
++ L K+V P L D +++V S +IG G++++E+RE
Sbjct 103 TV-------LNILKDVLPRLEDNFSERDPCEVRMLVHQSHAGALIGRNGSKIKELREKCS 155
Query 170 TRITVDRNSSP-CPERIIAVHG 190
R+ + +P +R++ G
Sbjct 156 ARLKIFTGCAPGSTDRVLITSG 177
> Hs20539507
Length=579
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 63/122 (51%), Gaps = 5/122 (4%)
Query 62 LIDPALA-NSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGI 120
L PAL+ ++IGK+ I +++ G S+ + ++ P++ R+++++G E+
Sbjct 410 LFIPALSVGAIIGKQGQHIKQLSRFAGASIKIAPAEA--PDAKVRMVIITGPPEAQFKAQ 467
Query 121 IALLERCKEVHPNLPYDQ--MYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNS 178
+ + KE + P ++ + A + VP+ VIG GG V E++ + + V R+
Sbjct 468 GRIYGKIKEENFVSPKEEVKLEAHIRVPSFAAGRVIGKGGKTVNELQNLSSAEVVVPRDQ 527
Query 179 SP 180
+P
Sbjct 528 TP 529
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 35/151 (23%), Positives = 72/151 (47%), Gaps = 18/151 (11%)
Query 48 ERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRIL 107
++P LP ++LL+ ++IGKE ATI + ++ + V +N + ++ +
Sbjct 191 QKPCDLP----LRLLVPTQFVGAIIGKEGATIRNITKQTQSKIDVHRKENA--GAAEKSI 244
Query 108 LMSGSTESISTGIIALLERCKEVHPNLPY-DQMYAKLVVPTSVCPTVIGPGGTRVREIRE 166
+ + E S ++LE + ++ + +++ K++ + +IG G +++I +
Sbjct 245 TILSTPEGTSAACKSILEIMHKEAQDIKFTEEIPLKILAHNNFVGRLIGKEGRNLKKIEQ 304
Query 167 ATLTRITVDRNSSPC-------PERIIAVHG 190
T T+IT+ SP PER I V G
Sbjct 305 DTDTKITI----SPLQELTLYNPERTITVKG 331
> CE28190
Length=557
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 45/113 (39%), Gaps = 14/113 (12%)
Query 68 ANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERC 127
A +IGK I R+A E G + N P S DRI ++ G+ + I + E
Sbjct 251 AGMIIGKGGEMIKRLAAETGTKIQFKPDTN--PNSEDRIAVIMGTRDQIYRATERITEIV 308
Query 128 KEVHPN--LPYDQMYAKLV----------VPTSVCPTVIGPGGTRVREIREAT 168
N P D+ A V VP C VIG GG +++I T
Sbjct 309 NRAIKNNGAPQDRGSAGTVLPGQSIFYMHVPAGKCGLVIGKGGENIKQIERET 361
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 39/161 (24%), Positives = 63/161 (39%), Gaps = 22/161 (13%)
Query 49 RPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILL 108
+P + PG ++LI +IGK TI V E+ G + + + L
Sbjct 130 QPGAAPGEVTEEMLIPADKIGLVIGKGGETIRIVQEQSGLRNCNVVQETTTATGQPKPLR 189
Query 109 MSGSTESIST-------------GIIALLERCKEVHPNLPYDQMYA--------KLVVPT 147
M GS +I T G LL+R P+ + Y +++VP
Sbjct 190 MIGSPAAIETAKALVHNIMNNTQGNAPLLQRAPH-QPSGQFGGGYGAQEAQAKGEVIVPR 248
Query 148 SVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAV 188
+IG GG ++ + T T+I +++P E IAV
Sbjct 249 LSAGMIIGKGGEMIKRLAAETGTKIQFKPDTNPNSEDRIAV 289
> 7302361
Length=499
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 45/93 (48%), Gaps = 9/93 (9%)
Query 57 FHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESI 116
F V+LLI +LA +IGK I + + GC + S N P+STDR++ G +
Sbjct 91 FDVRLLIHQSLAGCVIGKGGQKIKEIRDRIGCRFLKVFS-NVAPQSTDRVVQTVGKQSQV 149
Query 117 STGIIALLERCKE------VHPNLP--YDQMYA 141
+ ++ ++ +H P +D++YA
Sbjct 150 IEAVREVITLTRDTPIKGAIHNYDPMNFDRVYA 182
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 53/108 (49%), Gaps = 10/108 (9%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
V++LI ++A ++IGK I ++ + ++ V S+ PE T +I ST I T
Sbjct 21 VRILIPSSIAGAVIGKGGQHIQKMRTQYKATVSVDDSQG--PERTIQISADIESTLEIIT 78
Query 119 GIIALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIRE 166
++ E E +L++ S+ VIG GG +++EIR+
Sbjct 79 EMLKYFEERDE--------DFDVRLLIHQSLAGCVIGKGGQKIKEIRD 118
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 29/47 (61%), Gaps = 2/47 (4%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAV 188
++++P+S+ VIG GG ++++R T + TV + S PER I +
Sbjct 22 RILIPSSIAGAVIGKGGQHIQKMR--TQYKATVSVDDSQGPERTIQI 66
> Hs5729901
Length=579
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 35/151 (23%), Positives = 72/151 (47%), Gaps = 18/151 (11%)
Query 48 ERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRIL 107
++P LP ++LL+ ++IGKE ATI + ++ + V +N + ++ +
Sbjct 191 QKPCDLP----LRLLVPTQFVGAIIGKEGATIRNITKQTQSKIDVHRKENA--GAAEKSI 244
Query 108 LMSGSTESISTGIIALLERCKEVHPNLPY-DQMYAKLVVPTSVCPTVIGPGGTRVREIRE 166
+ + E S ++LE + ++ + +++ K++ + +IG G +++I +
Sbjct 245 TILSTPEGTSAACKSILEIMHKEAQDIKFTEEIPLKILAHNNFVGRLIGKEGRNLKKIEQ 304
Query 167 ATLTRITVDRNSSPC-------PERIIAVHG 190
T T+IT+ SP PER I V G
Sbjct 305 DTDTKITI----SPLQELTLYNPERTITVKG 331
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 62/119 (52%), Gaps = 5/119 (4%)
Query 65 PALA-NSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIAL 123
PAL+ ++IGK+ I +++ G S+ + ++ P++ R+++++G E+ +
Sbjct 413 PALSVGAIIGKQGQHIKQLSRFAGASIKIAPAEA--PDAKVRMVIITGPPEAQFKAQGRI 470
Query 124 LERCKEVHPNLPYDQ--MYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSP 180
+ KE + P ++ + A + VP+ VIG GG V E++ + + V R+ +P
Sbjct 471 YGKIKEENFVSPKEEVKLEAHIRVPSFAAGRVIGKGGKTVNELQNLSSAEVVVPRDQTP 529
> YBL032w
Length=381
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 34/141 (24%), Positives = 65/141 (46%), Gaps = 24/141 (17%)
Query 58 HVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
+V+L++ + +S+IGK ATI + + G + +++SK+ P S +RI+ + G SI+
Sbjct 158 YVRLIVANSHISSIIGKAGATIKSLINKHG--VKIVASKDFLPASDERIIEIQGFPGSIT 215
Query 118 TGIIALLERC-KEVHPNLPYDQMY---------------------AKLVVPTSVCPTVIG 155
+I + E +V ++ Y +L +P +IG
Sbjct 216 NVLIEISEIILSDVDVRFSTERSYFPHLKKSSGEPTSPSTSSNTRIELKIPELYVGAIIG 275
Query 156 PGGTRVREIREATLTRITVDR 176
G R++ ++ T T I V+R
Sbjct 276 RGMNRIKNLKTFTKTNIVVER 296
> Hs17402900
Length=644
Score = 39.7 bits (91), Expect = 0.004, Method: Composition-based stats.
Identities = 33/151 (21%), Positives = 65/151 (43%), Gaps = 19/151 (12%)
Query 31 LQEGMNMEALKEDPLEDERPVSLPGPFHVK---------LLIDPALANSLIGKESATITR 81
L++G +A K P D LP P H + + + +IG+ I+R
Sbjct 67 LEDGDQPDAKKVAPQNDSFGTQLP-PMHQQQSRSVMTEEYKVPDGMVGFIIGRGGEQISR 125
Query 82 VAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKEVHPNLPYDQ--- 138
+ +E GC + + PE R +++G+ ES+ + L + ++ P +
Sbjct 126 IQQESGCKIQIAPDSGGLPE---RSCMLTGTPESVQSAKRLLDQIVEKGRPAPGFHHGDG 182
Query 139 ---MYAKLVVPTSVCPTVIGPGGTRVREIRE 166
++++P S VIG GG +++++E
Sbjct 183 PGNAVQEIMIPASKAGLVIGKGGETIKQLQE 213
Score = 30.4 bits (67), Expect = 2.0, Method: Composition-based stats.
Identities = 33/148 (22%), Positives = 59/148 (39%), Gaps = 14/148 (9%)
Query 54 PGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPEST--DRILLMSG 111
PG +++I + A +IGK TI ++ E G M ++ P++T D+ L ++G
Sbjct 183 PGNAVQEIMIPASKAGLVIGKGGETIKQLQERAGVKMVMIQDG---PQNTGADKPLRITG 239
Query 112 STESISTGIIALLERCKEVHPNLPYDQMYAKLV---------VPTSVCPTVIGPGGTRVR 162
+ +LE ++ Y + +P VIG G ++
Sbjct 240 DPYKVQQAKEMVLELIRDQGGFREVRNEYGSRIGGNEGIDVPIPRFAVGIVIGRNGEMIK 299
Query 163 EIREATLTRITVDRNSSPCPERIIAVHG 190
+I+ RI + PERI + G
Sbjct 300 KIQNDAGVRIQFKPDDGTTPERIAQITG 327
Score = 30.0 bits (66), Expect = 3.2, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 143 LVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSP 180
+VPT +IG GG ++ I + + RI + RN P
Sbjct 381 FIVPTGKTGLIIGKGGETIKSISQQSGARIELQRNPPP 418
> Hs14670371
Length=369
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 50/96 (52%), Gaps = 8/96 (8%)
Query 104 DRILLMSGSTESISTGIIALLERCKE---VHP----NLPYDQMYAKLVVPTSVCPTVIGP 156
+RI ++GST ++ + + + E P N+ + +LV+P S C ++IG
Sbjct 26 ERITTITGSTAAVFHAVSMIAFKLDEDLCAAPANGGNVSRPPVTLRLVIPASQCGSLIGK 85
Query 157 GGTRVREIREATLTRITVDRNSSP-CPERIIAVHGM 191
GT+++EIRE T ++ V + P ER + V G+
Sbjct 86 AGTKIKEIRETTGAQVQVAGDLLPNSTERAVTVSGV 121
> HsM5729882
Length=577
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 29/124 (23%), Positives = 59/124 (47%), Gaps = 4/124 (3%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
V++ I ++IGK+ I +++ S+ + + P+S R+++++G E+
Sbjct 408 VQVFIPAQAVGAIIGKKGQHIKQLSRFASASIKIAPPET--PDSKVRMVIITGPPEAQFK 465
Query 119 GIIALLERCKEVHPNLPYDQ--MYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDR 176
+ + KE + P ++ + + VP S VIG GG V E++ T + V R
Sbjct 466 AQGRIYGKLKEENFFGPKEEVKLETHIRVPASAAGRVIGKGGKTVNELQNLTAAEVVVPR 525
Query 177 NSSP 180
+ +P
Sbjct 526 DQTP 529
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 32/136 (23%), Positives = 63/136 (46%), Gaps = 6/136 (4%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
++LL+ ++IGKE ATI + ++ + V +N + ++ + + + E S+
Sbjct 198 LRLLVPTQYVGAIIGKEGATIRNITKQTQSKIDVHRKEN--AGAAEKAISVHSTPEGCSS 255
Query 119 GIIALLE-RCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRN 177
+LE KE D++ K++ + +IG G ++++ + T T+IT+
Sbjct 256 ACKMILEIMHKEAKDTKTADEVPLKILAHNNFVGRLIGKEGRNLKKVEQDTETKITISSL 315
Query 178 SSPC---PERIIAVHG 190
PER I V G
Sbjct 316 QDLTLYNPERTITVKG 331
> Hs21361352
Length=577
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 29/124 (23%), Positives = 59/124 (47%), Gaps = 4/124 (3%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
V++ I ++IGK+ I +++ S+ + + P+S R+++++G E+
Sbjct 408 VQVFIPAQAVGAIIGKKGQHIKQLSRFASASIKIAPPET--PDSKVRMVIITGPPEAQFK 465
Query 119 GIIALLERCKEVHPNLPYDQ--MYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDR 176
+ + KE + P ++ + + VP S VIG GG V E++ T + V R
Sbjct 466 AQGRIYGKLKEENFFGPKEEVKLETHIRVPASAAGRVIGKGGKTVNELQNLTAAEVVVPR 525
Query 177 NSSP 180
+ +P
Sbjct 526 DQTP 529
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 32/136 (23%), Positives = 62/136 (45%), Gaps = 6/136 (4%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
++LL+ ++IGKE ATI + ++ + V +N + ++ + + + E S+
Sbjct 198 LRLLVPTQYVGAIIGKEGATIRNITKQTQSKIDVHRKEN--AGAAEKAISVHSTPEGCSS 255
Query 119 GIIALLE-RCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRN 177
+LE KE D++ K + + +IG G ++++ + T T+IT+
Sbjct 256 ACKMILEIMHKEAKDTKTADEVPLKTLAHNNFVGRLIGKEGRNLKKVEQDTETKITISSL 315
Query 178 SSPC---PERIIAVHG 190
PER I V G
Sbjct 316 QDLTLYNPERTITVKG 331
> 7292558
Length=558
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 37/157 (23%), Positives = 75/157 (47%), Gaps = 23/157 (14%)
Query 53 LPGP-----FHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRIL 107
+PGP F +++L+ + ++IG++ +TI + ++ + V +N S ++ +
Sbjct 69 MPGPGRQADFPLRILVQSEMVGAIIGRQGSTIRTITQQSRARVDVHRKENV--GSLEKSI 126
Query 108 LMSGSTESISTGIIALLERCK---------EVHPNLPYDQMYAKLVVPTSVCPTVIGPGG 158
+ G+ E+ + +LE + E+ P ++ K++ ++ +IG G
Sbjct 127 TIYGNPENCTNACKRILEVMQQEAISTNKGELSPEC--SEICLKILAHNNLIGRIIGKSG 184
Query 159 TRVREIREATLTRITV----DRNSSPCPERIIAVHGM 191
++ I + T T+ITV D NS ERII V G+
Sbjct 185 NTIKRIMQDTDTKITVSSINDINSFNL-ERIITVKGL 220
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 25/112 (22%), Positives = 48/112 (42%), Gaps = 4/112 (3%)
Query 61 LLIDPALANSLIGKESATITRVAEECGCSMHV--LSSKNKFPESTDRILLMSGSTESIST 118
L I ++IG + I + S+ + L + + T+R + + G+ E
Sbjct 307 LYIPNNAVGAIIGTRGSHIRSIMRFSNASLKIAPLDADKPLDQQTERKVTIVGTPEGQWK 366
Query 119 GIIALLERCKEVHPNLPYD--QMYAKLVVPTSVCPTVIGPGGTRVREIREAT 168
+ E+ +E D ++ +L+V +S +IG GG VRE++ T
Sbjct 367 AQYMIFEKMREEGFMCGTDDVRLTVELLVASSQVGRIIGKGGQNVRELQRVT 418
> Hs4885409
Length=1268
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 46/109 (42%), Gaps = 10/109 (9%)
Query 58 HVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
+V++ ID LIGK A I R+ ++ S+ + K ++ + G + +
Sbjct 437 YVEINIDHKFHRHLIGKSGANINRIKDQYKVSVRIPPDSEK-----SNLIRIEGDPQGVQ 491
Query 118 TGIIALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIRE 166
LLE + D L++ T+IG G R+REIR+
Sbjct 492 QAKRELLELASRMENERTKD-----LIIEQRFHRTIIGQKGERIREIRD 535
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 56/118 (47%), Gaps = 16/118 (13%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIST 118
V++ I L NSLIG + I + EECG +H+ FP + SGS +
Sbjct 656 VEVSIPAKLHNSLIGTKGRLIRSIMEECG-GVHI-----HFP------VEGSGSDTVVIR 703
Query 119 GIIALLERCKEVHPNLPYDQMYAKLVVPTSVCPT----VIGPGGTRVREIREATLTRI 172
G + +E+ K+ +L ++ V P +IG GG ++R++R++T R+
Sbjct 704 GPSSDVEKAKKQLLHLAEEKQTKSFTVDIRAKPEYHKFLIGKGGGKIRKVRDSTGARV 761
Score = 32.7 bits (73), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 51/107 (47%), Gaps = 15/107 (14%)
Query 61 LLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPES---TDRILLMSGSTESIS 117
+L+DP + + + +AEE G M FP S +D++ L G+ + +
Sbjct 805 MLVDPKHHRHFVIRRGQVLREIAEEYGGVM------VSFPRSGTQSDKVTL-KGAKDCVE 857
Query 118 TGIIALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREI 164
A +R +E+ +L Q+ + +P +V+GP G+R+++I
Sbjct 858 ----AAKKRIQEIIEDLEA-QVTLECAIPQKFHRSVMGPKGSRIQQI 899
Score = 32.3 bits (72), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 30/139 (21%), Positives = 59/139 (42%), Gaps = 9/139 (6%)
Query 56 PFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTES 115
P +++ + L +IG++ + I ++ +E ++HV + PE I+ ++G +
Sbjct 972 PVTIEVEVPFDLHRYVIGQKGSGIRKMMDEFEVNIHVPA-----PELQSDIIAITGLAAN 1026
Query 116 ISTGIIALLERCKEVHP---NLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRI 172
+ LLER KE+ + + V P +IG G + +IR I
Sbjct 1027 LDRAKAGLLERVKELQAEQEDRALRSFKLSVTVDPKYHPKIIGRKGAVITQIRLEHDVNI 1086
Query 173 TV-DRNSSPCPERIIAVHG 190
D++ P+ I + G
Sbjct 1087 QFPDKDDGNQPQDQITITG 1105
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 34/160 (21%), Positives = 71/160 (44%), Gaps = 21/160 (13%)
Query 26 AETGVLQEGMNMEALKEDPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEE 85
A+ G+L+ ++A +ED R + F + + +DP +IG++ A IT++ E
Sbjct 1030 AKAGLLERVKELQAEQED-----RALR---SFKLSVTVDPKYHPKIIGRKGAVITQIRLE 1081
Query 86 CGCSMHVLSSKNKFPESTDRILL--MSGSTESISTGIIALLERCKE-VHPNLPYDQMYAK 142
++ K+ + D+I + +TE+ I+ ++ ++ V ++P D
Sbjct 1082 HDVNIQ-FPDKDDGNQPQDQITITGYEKNTEAARDAILRIVGELEQMVSEDVPLDH---- 1136
Query 143 LVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCP 182
V +IG G +R+I + I ++ +P P
Sbjct 1137 -----RVHARIIGARGKAIRKIMDEFKVDIRFPQSGAPDP 1171
> 7302610
Length=1268
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 46/179 (25%), Positives = 74/179 (41%), Gaps = 24/179 (13%)
Query 19 KLKMPAWAETGVLQEGMNMEALKEDPLEDERPVS-----LPGPFHVKLLIDPALANSLIG 73
K+ +PA +T E + + KE+ LE + + L ++ I P NS+IG
Sbjct 596 KIDLPAEGDTN---EVIVITGKKENVLEAKERIQKIQNELSDIVTEEVQIPPKYYNSIIG 652
Query 74 KESATITRVAEEC-GCSMHVLSSKNKFPES---TDRILLMSGSTESISTGIIALLERCKE 129
I+ + EEC G S+ KFP S +D++ + G + + + LLE E
Sbjct 653 TGGKLISSIMEECGGVSI-------KFPNSDSKSDKVTI-RGPKDDVEKAKVQLLELANE 704
Query 130 VHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAV 188
A++ +IG G +R+IR+AT RI N E I +
Sbjct 705 RQ----LASFTAEVRAKQQHHKFLIGKNGASIRKIRDATGARIIFPSNEDTDKEVITII 759
Score = 33.5 bits (75), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 63/149 (42%), Gaps = 27/149 (18%)
Query 29 GVLQEGMNMEALKEDPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGC 88
GV Q + ++ K D LE+E+ + +ID L S+IG + I V +
Sbjct 471 GVRQAQLELQE-KIDKLENEKSKDV--------IIDRRLHRSIIGAKGEKIREVKDRYRQ 521
Query 89 SMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKEVHPN-----LPYDQMYAKL 143
+ + P+ I+ + G E + LL+ KE+ + +P + + K
Sbjct 522 VTITIPT----PQENTDIVKLRGPKEDVDKCHKDLLKLVKEIQESSHIIEVPIFKQFHKF 577
Query 144 VVPTSVCPTVIGPGGTRVREIREATLTRI 172
V IG GG +++IR+ T T+I
Sbjct 578 V---------IGKGGANIKKIRDETQTKI 597
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 25/105 (23%), Positives = 50/105 (47%), Gaps = 15/105 (14%)
Query 63 IDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPE---STDRILLMSGSTESISTG 119
+DP + K + R++EECG M FP ++D++ + G+ + I
Sbjct 789 VDPKHHKHFVAKRGFILHRISEECGGVMI------SFPRVGINSDKVTI-KGAKDCIE-- 839
Query 120 IIALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREI 164
A +R +E+ +L Q ++V+P T++G G +V+++
Sbjct 840 --AARQRIEEIVADLEA-QTTIEVVIPQRHHRTIMGARGFKVQQV 881
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 20/114 (17%), Positives = 52/114 (45%), Gaps = 15/114 (13%)
Query 61 LLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGI 120
+ ++P+ +IGK A + R+ +E ++++ + + + + G E +
Sbjct 423 MTVNPSYYKHIIGKAGANVNRLKDELKVNINIEEREGQ------NNIRIEGPKEGVRQAQ 476
Query 121 IALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREI----REATLT 170
+ L E+ ++ D +++ + ++IG G ++RE+ R+ T+T
Sbjct 477 LELQEKIDKLENEKSKD-----VIIDRRLHRSIIGAKGEKIREVKDRYRQVTIT 525
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITV 174
++ VP ++G GG R+REI T TRI +
Sbjct 139 QVTVPREHFRVILGKGGQRLREIERVTATRINI 171
> CE26796
Length=618
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 60/125 (48%), Gaps = 9/125 (7%)
Query 70 SLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESI---STGIIALLER 126
+++G + I ++++E G + L + P+ +R L + G+ + + I A++E
Sbjct 236 AIMGLQGKNIKKLSDETGTKIQFLPDDD--PKLMERSLAIIGNKNKVYVCAQLIKAIVEA 293
Query 127 CKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCP-ERI 185
E N P Y +V+P S C VIG GG +++I + + + R+ + P E+
Sbjct 294 NSEAA-NAPVVLFY--MVIPASKCGLVIGRGGETIKQINQESGAHCELSRDPNTNPIEKT 350
Query 186 IAVHG 190
+ G
Sbjct 351 FVIRG 355
> CE26797
Length=641
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 60/125 (48%), Gaps = 9/125 (7%)
Query 70 SLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESI---STGIIALLER 126
+++G + I ++++E G + L + P+ +R L + G+ + + I A++E
Sbjct 259 AIMGLQGKNIKKLSDETGTKIQFLPDDD--PKLMERSLAIIGNKNKVYVCAQLIKAIVEA 316
Query 127 CKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCP-ERI 185
E N P Y +V+P S C VIG GG +++I + + + R+ + P E+
Sbjct 317 NSEA-ANAPVVLFY--MVIPASKCGLVIGRGGETIKQINQESGAHCELSRDPNTNPIEKT 373
Query 186 IAVHG 190
+ G
Sbjct 374 FVIRG 378
> 7302869
Length=797
Score = 37.4 bits (85), Expect = 0.018, Method: Composition-based stats.
Identities = 34/143 (23%), Positives = 56/143 (39%), Gaps = 33/143 (23%)
Query 71 LIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTG---IIALLERC 127
+IGK I ++ ECGC + + KN E DR ++ G+ + + I L+E
Sbjct 330 VIGKGGDMIRKIQTECGCKLQFIQGKND--EMGDRRCVIQGTRQQVDDAKRTIDGLIENV 387
Query 128 KE---------------------VHPNLPYDQMYAK-------LVVPTSVCPTVIGPGGT 159
+ + Y +A+ +VP S C VIG GG
Sbjct 388 MQRNGMNRNGNGGGGGPGGDSGNSNYGYGYGVNHAQGGREEITFLVPASKCGIVIGRGGE 447
Query 160 RVREIREATLTRITVDRNSSPCP 182
++ I + + +DRN+S P
Sbjct 448 TIKLINQQSGAHTEMDRNASNPP 470
Score = 28.9 bits (63), Expect = 5.7, Method: Composition-based stats.
Identities = 24/124 (19%), Positives = 56/124 (45%), Gaps = 22/124 (17%)
Query 66 ALANSLIGKESA-TITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTG----- 119
++A + +G+ S TIT + E G + V+ + DR++++ G ++++ G
Sbjct 131 SVAGAFMGRSSNDTITHIQAESGVKVQVMQDQ-------DRVIMLRGQRDTVTKGREMIQ 183
Query 120 ---------IIALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLT 170
+ +L P Y ++++P + VIG GG +++++E T
Sbjct 184 NMANRAGGGQVEVLLTINMPPPGPSGYPPYQEIMIPGAKVGLVIGKGGDTIKQLQEKTGA 243
Query 171 RITV 174
++ +
Sbjct 244 KMII 247
> Hs18570703
Length=416
Score = 37.0 bits (84), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 31/151 (20%), Positives = 70/151 (46%), Gaps = 19/151 (12%)
Query 37 MEALKEDPLEDERPVSLPGPFHVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSK 96
+E+++++ E + +P +K++ L LIGKE + + E G + + S +
Sbjct 172 LESMQKEADETKLAEEIP----LKIVAHNGLFGRLIGKEGRNVKKNEHETGTKITISSLQ 227
Query 97 NKFPESTDRILLMSGSTESISTG---IIALLERCKEVHPN-----LPYDQMYAK-----L 143
+ + +R + + G+ E+ ++ I+ L ++P+ P+ Y + L
Sbjct 228 DLSICNPERTITVKGTVEAYASAEMEIMKKLHTFSSLYPHHQFGPFPHQHSYQEQEIVNL 287
Query 144 VVPTSVCPTVIGPGGTRVREIREATLTRITV 174
+PT T+IG G ++++ A L R ++
Sbjct 288 FIPTQAVGTIIGKKGAHIKQL--ARLARASI 316
> Hs5803195
Length=606
Score = 36.6 bits (83), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 58/122 (47%), Gaps = 12/122 (9%)
Query 71 LIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKEV 130
+IG++ A I ++ ++ G + V + +R+LL+SG + A+ + E
Sbjct 67 IIGRQGANIKQLRKQTGARIDVDTEDV----GDERVLLISGFPVQVCKAKAAIHQILTE- 121
Query 131 HPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNS--SPCPERIIAV 188
N P + +L VP +IG GG +R I +A+ +IT D+ S + R+I +
Sbjct 122 --NTPVSE---QLSVPQRSVGRIIGRGGETIRSICKASGAKITCDKESEGTLLLSRLIKI 176
Query 189 HG 190
G
Sbjct 177 SG 178
> At1g51580
Length=621
Score = 36.2 bits (82), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 46/174 (26%), Positives = 77/174 (44%), Gaps = 27/174 (15%)
Query 23 PAWAETGVLQEGMNME--ALKEDPLEDERPVS-LP---GPFH---------VKLLIDPAL 67
P W E Q G E +++ P D P + +P GPF+ +LL
Sbjct 227 PTWDECPFPQPGYPPEYHSMEYHPQWDHPPPNPMPEDVGPFNRPVVEEEVAFRLLCPADK 286
Query 68 ANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESI-------STGI 120
SLIGK A + + E G S+ V + +S +RI+++S + E++ G+
Sbjct 287 VGSLIGKGGAVVRALQNESGASIKVSDPTH---DSEERIIVIS-ARENLERRHSLAQDGV 342
Query 121 IALLERCKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITV 174
+ + R E+ P + A+L+V + ++G GG + E+R AT I V
Sbjct 343 MRVHNRIVEIGFE-PSAAVVARLLVHSPYIGRLLGKGGHLISEMRRATGASIRV 395
Score = 28.5 bits (62), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 137 DQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITV-DRNSSPCPERIIAV 188
+ ++ +L+ P + +IG GG+ +R ++ T ++I V D P ER++ +
Sbjct 17 ESVHFRLLCPATRTGAIIGKGGSVIRHLQSVTGSKIRVIDDIPVPSEERVVLI 69
> Hs20475146
Length=344
Score = 36.2 bits (82), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 39/166 (23%), Positives = 73/166 (43%), Gaps = 21/166 (12%)
Query 33 EGMNMEALKEDPLEDERPVSL--------PGPFH--------VKLLIDPALANSLIGKES 76
E M EA E+ L +E P+ + P P H V L I ++I K
Sbjct 162 EIMQKEA-DENKLAEEIPLKILAHNGLVGPFPHHHSYPEQEVVNLFIPTQAVGAIIRKRG 220
Query 77 ATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKEVHPNLPY 136
A I ++A S+ + ++ P+ +R+++++ E+ + + KE + P
Sbjct 221 AHIKQLARFATASIKIALAEG--PDVNERMVIITRPPEAQFKAQGRIFGKLKEENFFNPK 278
Query 137 DQ--MYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSP 180
++ + A++ VP+S VIG G + E++ T + V R+ P
Sbjct 279 EEVKLEARIRVPSSTAGRVIGKGVNTLNELQNLTSAEVIVPRDQRP 324
> Hs14165437
Length=464
Score = 36.2 bits (82), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 44/92 (47%), Gaps = 12/92 (13%)
Query 58 HVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
++LLI +LA +IG + A I + E ++ + + P STDR++L+ G + +
Sbjct 146 ELRLLIHQSLAGGIIGVKGAKIKELRENTQTTIKLF--QECCPHSTDRVVLIGGKPDRVV 203
Query 118 TGIIALLE---------RCKEVHPNLPYDQMY 140
I +L+ R + PN YD+ Y
Sbjct 204 ECIKIILDLISESPIKGRAQPYDPNF-YDETY 234
Score = 35.0 bits (79), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHG 190
++ +P + ++IG GG R+++IR + I +D +RII + G
Sbjct 391 QVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITG 439
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 39/159 (24%), Positives = 68/159 (42%), Gaps = 31/159 (19%)
Query 58 HVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
+++L+ A ++IGK I + + S+ V S PE RIL +S E+I
Sbjct 44 ELRILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSG--PE---RILSISADIETIG 98
Query 118 ---TGIIALLERCKEV-----HPNLPYD---------QMYA--------KLVVPTSVCPT 152
II LE ++ LP + Q Y +L++ S+
Sbjct 99 EILKKIIPTLEEGLQLPSPTATSQLPLESDAVECLNYQHYKGSDFDCELRLLIHQSLAGG 158
Query 153 VIGPGGTRVREIREATLTRITVDRNSSP-CPERIIAVHG 190
+IG G +++E+RE T T I + + P +R++ + G
Sbjct 159 IIGVKGAKIKELRENTQTTIKLFQECCPHSTDRVVLIGG 197
> Hs14165435
Length=463
Score = 36.2 bits (82), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 44/92 (47%), Gaps = 12/92 (13%)
Query 58 HVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
++LLI +LA +IG + A I + E ++ + + P STDR++L+ G + +
Sbjct 146 ELRLLIHQSLAGGIIGVKGAKIKELRENTQTTIKLF--QECCPHSTDRVVLIGGKPDRVV 203
Query 118 TGIIALLE---------RCKEVHPNLPYDQMY 140
I +L+ R + PN YD+ Y
Sbjct 204 ECIKIILDLISESPIKGRAQPYDPNF-YDETY 234
Score = 35.0 bits (79), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHG 190
++ +P + ++IG GG R+++IR + I +D +RII + G
Sbjct 391 QVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITG 439
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 39/159 (24%), Positives = 68/159 (42%), Gaps = 31/159 (19%)
Query 58 HVKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESIS 117
+++L+ A ++IGK I + + S+ V S PE RIL +S E+I
Sbjct 44 ELRILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSG--PE---RILSISADIETIG 98
Query 118 ---TGIIALLERCKEV-----HPNLPYD---------QMYA--------KLVVPTSVCPT 152
II LE ++ LP + Q Y +L++ S+
Sbjct 99 EILKKIIPTLEEGLQLPSPTATSQLPLESDAVECLNYQHYKGSDFDCELRLLIHQSLAGG 158
Query 153 VIGPGGTRVREIREATLTRITVDRNSSP-CPERIIAVHG 190
+IG G +++E+RE T T I + + P +R++ + G
Sbjct 159 IIGVKGAKIKELRENTQTTIKLFQECCPHSTDRVVLIGG 197
> At5g64390
Length=833
Score = 36.2 bits (82), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 32/127 (25%), Positives = 54/127 (42%), Gaps = 14/127 (11%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGS------ 112
K+L A +IG + + E G ++V N + +R++ ++ S
Sbjct 454 FKILCSTENAGGVIGTGGKVVRMLHSETGAFINV---GNTLDDCEERLIAVTASENPECQ 510
Query 113 TESISTGIIALLERCKEVHPNL-----PYDQMYAKLVVPTSVCPTVIGPGGTRVREIREA 167
+ I+ + R E+ N P + A+LVVPTS V+G GG V E+R+
Sbjct 511 SSPAQKAIMLIFSRLFELATNKILDNGPRSSITARLVVPTSQIGCVLGKGGVIVSEMRKT 570
Query 168 TLTRITV 174
T I +
Sbjct 571 TGAAIQI 577
Score = 32.0 bits (71), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 0/55 (0%)
Query 134 LPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAV 188
+P +L+ P S VIG G +++++++T +I V+ S P+R+I +
Sbjct 42 VPVGHAAFRLLCPLSHVGAVIGKSGNVIKQLQQSTGAKIRVEEPPSGSPDRVITI 96
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAV 188
K++ T VIG GG VR + T I V C ER+IAV
Sbjct 455 KILCSTENAGGVIGTGGKVVRMLHSETGAFINVGNTLDDCEERLIAV 501
> Hs7662254
Length=891
Score = 35.8 bits (81), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Query 135 PYDQMYAKLVVPTSVCPT-VIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHG 190
P MY+ LV P ++C ++ P G R RE L + D N S E +A HG
Sbjct 621 PIKDMYSGLVGPLAICQKGILEPHGGRSDMDREFALLFLIFDENKSWYLEENVATHG 677
> Hs21166385
Length=1158
Score = 35.8 bits (81), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Query 135 PYDQMYAKLVVPTSVCPT-VIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHG 190
P MY+ LV P ++C ++ P G R RE L + D N S E +A HG
Sbjct 888 PIKDMYSGLVGPLAICQKGILEPHGGRSDMDREFALLFLIFDENKSWYLEENVATHG 944
> At4g18375
Length=606
Score = 35.8 bits (81), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 55/110 (50%), Gaps = 7/110 (6%)
Query 71 LIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTES----ISTGIIALLER 126
+IGK +TI R+ E G + V S+ K + D +++ +TES S + A+L
Sbjct 326 VIGKGGSTIKRIREASGSCIEVNDSRTKCGD--DECVIIVTATESPDDMKSMAVEAVL-L 382
Query 127 CKEVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDR 176
+E + + + +L+V + V VIG G+ + EIR+ T I + +
Sbjct 383 LQEYINDEDAENVKMQLLVSSKVIGCVIGKSGSVINEIRKRTNANICISK 432
Score = 35.0 bits (79), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 51/106 (48%), Gaps = 12/106 (11%)
Query 60 KLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPEST-----DRILLMSGSTE 114
+LL+ + ++SLIGK I R+ S+ V+S P D ++++SG E
Sbjct 142 RLLVPFSQSSSLIGKAGENIKRIRRRTRASVKVVSKDVSDPSHVCAMEYDNVVVISGEPE 201
Query 115 SISTGIIALLERCKEVHP--NLPYDQM-----YAKLVVPTSVCPTV 153
S+ + A+ +++P N+P D A ++VP+ + +V
Sbjct 202 SVKQALFAVSAIMYKINPRENIPLDSTSQDVPAASVIVPSDLSNSV 247
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Query 131 HPNLPYDQMYA-KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVH 189
H + D++ +++ P V VIG G + IR T +I V C +R+I ++
Sbjct 27 HDKINRDELVVYRILCPIDVVGGVIGKSGKVINAIRHNTKAKIKVFDQLHGCSQRVITIY 86
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 137 DQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPE 183
+++ K++ P VIG GG+ ++ IREA+ + I V+ + + C +
Sbjct 310 EELVFKVLCPLCNIMRVIGKGGSTIKRIREASGSCIEVNDSRTKCGD 356
> CE03509
Length=854
Score = 35.4 bits (80), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 59/129 (45%), Gaps = 8/129 (6%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHV--LSSKNK-FPESTDRILLMSGSTES 115
++ +++ +IG +TI +A C + LS K + + DRIL + G E
Sbjct 313 IRCVVEGKYHAVIIGPNGSTIKDIASSTRCRVDFVNLSKKERTVLGNNDRILTVHGVAEQ 372
Query 116 ISTGIIALLERCK----EVHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTR 171
+ + +L+ + + N+ D + ++ +C +IG G+ ++EI + T T
Sbjct 373 ATKAVARILDVIQSEAVKDDVNVGADTVL-RMRAHNQLCGRLIGKAGSSIKEIMQKTGTN 431
Query 172 ITVDRNSSP 180
ITV + P
Sbjct 432 ITVTKYIEP 440
> CE05266
Length=589
Score = 35.0 bits (79), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 34/146 (23%), Positives = 60/146 (41%), Gaps = 15/146 (10%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESI-- 116
+ + I P +IGK TI ++ E+ GC M +L N+ + L ++G + I
Sbjct 135 IDIAIPPNRCGLIIGKSGDTIRQLQEKSGCKM-ILVQDNQSVSDQSKPLRITGDPQKIEL 193
Query 117 STGIIALLERCKEVHPNLPYDQMY-----------AKLVVPTSVCPTVIGPGGTRVREIR 165
+ ++A + QM+ ++VVP S +IG G ++ +
Sbjct 194 AKQLVAEILNSGGDGNGGSGLQMHHAGGGGGASARGEVVVPRSSVGIIIGKQGDTIKRLA 253
Query 166 EATLTRITVDRNSSPC-PERIIAVHG 190
T T+I + P PER + G
Sbjct 254 METGTKIQFKPDDDPSTPERCAVIMG 279
Score = 32.0 bits (71), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 27/132 (20%), Positives = 54/132 (40%), Gaps = 14/132 (10%)
Query 71 LIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKE- 129
+IGK+ TI R+A E G + + P + +R ++ G+ + I + E K+
Sbjct 241 IIGKQGDTIKRLAMETGTKIQFKPDDD--PSTPERCAVIMGTRDQIYRATERITELVKKS 298
Query 130 ----------VHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRN-S 178
+ + + VP + C VIG GG +++I + + R+ +
Sbjct 299 TMQQGGGGNVAGAMVSNEASTFYMSVPAAKCGLVIGKGGETIKQINSESGAHCELSRDPT 358
Query 179 SPCPERIIAVHG 190
E++ + G
Sbjct 359 GNADEKVFVIKG 370
> CE05267
Length=611
Score = 35.0 bits (79), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 34/146 (23%), Positives = 60/146 (41%), Gaps = 15/146 (10%)
Query 59 VKLLIDPALANSLIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESI-- 116
+ + I P +IGK TI ++ E+ GC M +L N+ + L ++G + I
Sbjct 157 IDIAIPPNRCGLIIGKSGDTIRQLQEKSGCKM-ILVQDNQSVSDQSKPLRITGDPQKIEL 215
Query 117 STGIIALLERCKEVHPNLPYDQMY-----------AKLVVPTSVCPTVIGPGGTRVREIR 165
+ ++A + QM+ ++VVP S +IG G ++ +
Sbjct 216 AKQLVAEILNSGGDGNGGSGLQMHHAGGGGGASARGEVVVPRSSVGIIIGKQGDTIKRLA 275
Query 166 EATLTRITVDRNSSPC-PERIIAVHG 190
T T+I + P PER + G
Sbjct 276 METGTKIQFKPDDDPSTPERCAVIMG 301
Score = 32.0 bits (71), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 27/132 (20%), Positives = 54/132 (40%), Gaps = 14/132 (10%)
Query 71 LIGKESATITRVAEECGCSMHVLSSKNKFPESTDRILLMSGSTESISTGIIALLERCKE- 129
+IGK+ TI R+A E G + + P + +R ++ G+ + I + E K+
Sbjct 263 IIGKQGDTIKRLAMETGTKIQFKPDDD--PSTPERCAVIMGTRDQIYRATERITELVKKS 320
Query 130 ----------VHPNLPYDQMYAKLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRN-S 178
+ + + VP + C VIG GG +++I + + R+ +
Sbjct 321 TMQQGGGGNVAGAMVSNEASTFYMSVPAAKCGLVIGKGGETIKQINSESGAHCELSRDPT 380
Query 179 SPCPERIIAVHG 190
E++ + G
Sbjct 381 GNADEKVFVIKG 392
> Hs19923832
Length=942
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHG 190
KL VP SV ++G GG + I++ T I VD+ ER+I + G
Sbjct 82 KLSVPASVVSRIMGRGGCNITAIQDVTGAHIDVDKQKDKNGERMITIRG 130
> 7301133
Length=2441
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 142 KLVVPTSVCPTVIGPGGTRVREIREATLTRITVDRNSSPCPERIIAVHGM 191
K+ VP + VIG GG+ + IR T I V++ ER I + G+
Sbjct 1480 KVQVPVNAISRVIGRGGSNINAIRATTGAHIEVEKQGKNQSERCITIKGL 1529
> Hs9966803
Length=424
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 38/70 (54%), Gaps = 7/70 (10%)
Query 104 DRILLMSGSTESISTGIIALLERCKE---VHP----NLPYDQMYAKLVVPTSVCPTVIGP 156
+RI ++GST ++ + + + E P N+ + +LV+P S C ++IG
Sbjct 26 ERITTITGSTAAVFHAVSMIAFKLDEDLCAAPANGGNVSRPPVTLRLVIPASQCGSLIGK 85
Query 157 GGTRVREIRE 166
GT+++EIRE
Sbjct 86 AGTKIKEIRE 95
Lambda K H
0.316 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3202455494
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40