bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3559_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
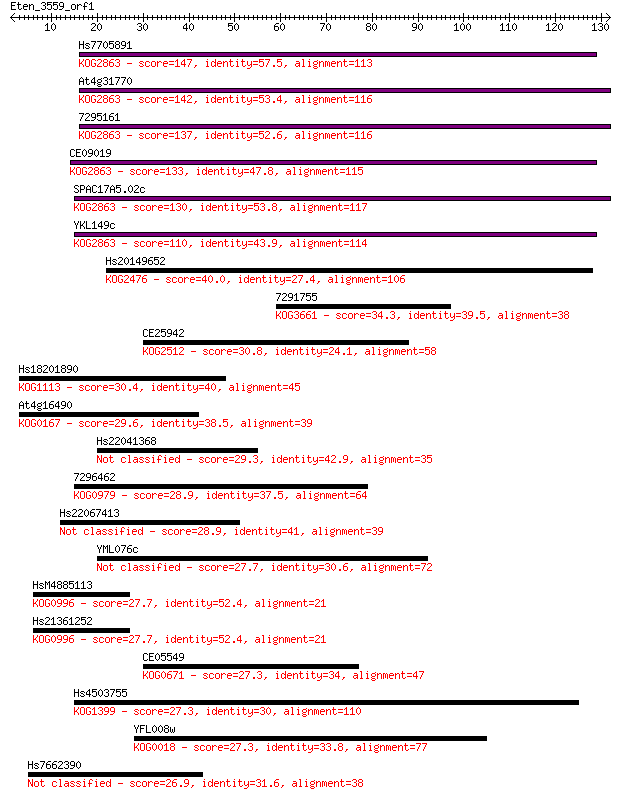
Hs7705891 147 5e-36
At4g31770 142 1e-34
7295161 137 4e-33
CE09019 133 7e-32
SPAC17A5.02c 130 8e-31
YKL149c 110 8e-25
Hs20149652 40.0 0.001
7291755 34.3 0.062
CE25942 30.8 0.71
Hs18201890 30.4 0.87
At4g16490 29.6 1.4
Hs22041368 29.3 1.8
7296462 28.9 2.4
Hs22067413 28.9 2.5
YML076c 27.7 5.3
HsM4885113 27.7 6.6
Hs21361252 27.7 6.6
CE05549 27.3 7.3
Hs4503755 27.3 7.5
YFL008w 27.3 7.6
Hs7662390 26.9 9.8
> Hs7705891
Length=544
Score = 147 bits (371), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 65/113 (57%), Positives = 87/113 (76%), Gaps = 0/113 (0%)
Query 16 MKVAINGCCHGELDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKYKK 75
M+VA+ GCCHGELD +YE L E + VDLL+CCGDFQ++R+EADL+ PPKY+
Sbjct 1 MRVAVAGCCHGELDKIYETLALAERRGPGPVDLLLCCGDFQAVRNEADLRCMAVPPKYRH 60
Query 76 LKDFHLYFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNIFFLGYSGVV 128
++ F+ Y+ G K+AP LTL +GGNHEA N L++L YGGW+APNI++LG +GVV
Sbjct 61 MQTFYRYYSGEKKAPVLTLFIGGNHEASNHLQELPYGGWVAPNIYYLGLAGVV 113
> At4g31770
Length=401
Score = 142 bits (359), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 62/116 (53%), Positives = 85/116 (73%), Gaps = 0/116 (0%)
Query 16 MKVAINGCCHGELDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKYKK 75
MK+AI GC HG+LD +Y+ +Q E+ N KVDLL+CCGDFQ++R+E D+ + P KY++
Sbjct 1 MKIAIEGCMHGDLDNVYKTIQHYEQIHNTKVDLLLCCGDFQAVRNEKDMDSLNVPRKYRE 60
Query 76 LKDFHLYFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNIFFLGYSGVVNVG 131
+K F Y+ G + AP T+ +GGNHEA N L +LYYGGW A NI+FLG++GVV G
Sbjct 61 MKSFWKYYSGQEVAPIPTIFIGGNHEASNYLWELYYGGWAATNIYFLGFAGVVKFG 116
> 7295161
Length=534
Score = 137 bits (346), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 61/116 (52%), Positives = 84/116 (72%), Gaps = 0/116 (0%)
Query 16 MKVAINGCCHGELDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKYKK 75
MK+A+ GC HGEL+ +Y+ ++ +E+ K+DLL+CCGDFQS R+ DLQ P KY
Sbjct 1 MKIAVEGCAHGELERIYDTIEGIEKVGGTKIDLLLCCGDFQSTRNLEDLQTMAVPKKYLD 60
Query 76 LKDFHLYFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNIFFLGYSGVVNVG 131
+ F+ Y+ G AP LT+ +GGNHEA N L++L YGGW+APNI++LGY+GVVNV
Sbjct 61 MCSFYKYYSGELVAPVLTIFIGGNHEASNYLQELPYGGWVAPNIYYLGYAGVVNVN 116
> CE09019
Length=504
Score = 133 bits (335), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 55/115 (47%), Positives = 85/115 (73%), Gaps = 0/115 (0%)
Query 14 EKMKVAINGCCHGELDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKY 73
+K+++A+ GC HGE+DA+YE + +E+K K DLLICCGD+Q++R+ DL + PPKY
Sbjct 39 KKIRIAVVGCSHGEMDAIYETMTLIEQKNGYKFDLLICCGDYQAVRNYGDLPHMSIPPKY 98
Query 74 KKLKDFHLYFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNIFFLGYSGVV 128
+ L+ F+ Y+ G ++AP LTL +GGNHEA L +L GGW+APNI+++G++ +
Sbjct 99 RSLQTFYKYYSGEQKAPVLTLFIGGNHEASGYLCELPNGGWVAPNIYYMGFANCI 153
> SPAC17A5.02c
Length=466
Score = 130 bits (326), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 63/117 (53%), Positives = 80/117 (68%), Gaps = 6/117 (5%)
Query 15 KMKVAINGCCHGELDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKYK 74
KM+V + GCCHG LD LY + E KVDLLI GDFQ+LR+ +D PPK+K
Sbjct 15 KMRVGVQGCCHGILDNLYIL------AEKRKVDLLIIGGDFQALRNVSDYHGISMPPKFK 68
Query 75 KLKDFHLYFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNIFFLGYSGVVNVG 131
+L DF Y+ G +AP LT+ VGGNHEA N L +L YGGW+APNI+++G S V+NVG
Sbjct 69 RLGDFFNYYNGRNKAPILTIFVGGNHEASNYLDELPYGGWVAPNIYYMGRSSVINVG 125
> YKL149c
Length=405
Score = 110 bits (275), Expect = 8e-25, Method: Composition-based stats.
Identities = 50/114 (43%), Positives = 75/114 (65%), Gaps = 2/114 (1%)
Query 15 KMKVAINGCCHGELDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKYK 74
K+++A+ GCCHG+L+ +Y+ + ++ K I DLLI GDFQS+R D + PPKY+
Sbjct 3 KLRIAVQGCCHGQLNQIYKEVSRIHAKTPI--DLLIILGDFQSIRDGQDFKSIAIPPKYQ 60
Query 75 KLKDFHLYFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNIFFLGYSGVV 128
+L DF Y+ AP T+ +GGNHE+ L L +GG++A NIF++GYS V+
Sbjct 61 RLGDFISYYNNEIEAPVPTIFIGGNHESMRHLMLLPHGGYVAKNIFYMGYSNVI 114
> Hs20149652
Length=538
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 48/106 (45%), Gaps = 16/106 (15%)
Query 22 GCCHGELDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKYKKLKDFHL 81
G G+ D L+ +Q +++K DLL+C G+F +A+ +
Sbjct 12 GDVEGKFDILFNRVQAIQKKSG-NFDLLLCVGNFFGSTQDAE---------------WEE 55
Query 82 YFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNIFFLGYSGV 127
Y G+K+ P T ++G N++ Q G LA NI +LG G+
Sbjct 56 YKTGIKKVPIQTYVLGANNQETVKYFQDADGCELAENITYLGRKGI 101
> 7291755
Length=1423
Score = 34.3 bits (77), Expect = 0.062, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 59 RSEADLQYFHAPPKYKKLKDFHLYFRGLK-RAPRLTLLV 96
R + D ++ P ++K+K FHL+F G+K AP T+ V
Sbjct 409 RLQGDAKFVKTPSGFEKIKSFHLHFYGVKFEAPNQTIRV 447
> CE25942
Length=371
Score = 30.8 bits (68), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query 30 ALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYFHAPPKYKKLKDFHLYFRGLK 87
++ QQL ++ + + I C + + +D+ +FH +Y +LKD ++ GL+
Sbjct 149 TIFTSCQQLRTRDCTSIRISILCPTEPIVENSSDIHFFHLAMRYPELKD-QMHAVGLR 205
> Hs18201890
Length=572
Score = 30.4 bits (67), Expect = 0.87, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query 3 KKDLGSSGFLPEKMKVAINGCCHGELDALYEMLQQ-----LEEKENIKVD 47
K + GSS FL K+ G C GE+D L+ +++ +EE E + VD
Sbjct 165 KDEDGSSAFLDPHPKLLHKGSCFGEMDVLHASVRRSTIVCMEETEFLVVD 214
> At4g16490
Length=459
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 5/44 (11%)
Query 3 KKDLGSSGFLPEKMKVAINGCCHGELDALYEM-----LQQLEEK 41
K +G+ G +P + + +NG C G+ DAL + LQQ +E+
Sbjct 169 KGSIGACGAIPPLVSLLLNGSCRGKKDALTTLYKLCTLQQNKER 212
> Hs22041368
Length=927
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 5/40 (12%)
Query 20 INGCCHGELDALYEMLQQ-----LEEKENIKVDLLICCGD 54
++ C H + D LYE +Q+ + EKEN +V+L C D
Sbjct 176 VHTCMHRKRDELYEEMQKAIIISIVEKENSEVNLRTCQSD 215
> 7296462
Length=1345
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 10/74 (13%)
Query 15 KMKVAINGCCHGEL--DALYEMLQQLEEKENIKV-DLLICCGDFQ-------SLRSEADL 64
K K AI G CHGE+ + + ++ E ENI + +L DFQ S+ SEA
Sbjct 747 KRKSAIQGLCHGEIPTSSKFPFKKEFRELENIDLPELREAIHDFQARLECMKSVNSEAIS 806
Query 65 QYFHAPPKYKKLKD 78
Y + K+L++
Sbjct 807 SYQGLQNEVKQLEE 820
> Hs22067413
Length=212
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 4/43 (9%)
Query 12 LPEKMKVAINGCCHGELDAL----YEMLQQLEEKENIKVDLLI 50
LPE++ +A+ GCC G+ + E++ LE E I V+LL+
Sbjct 61 LPEEVALALGGCCCGDPSPVPKVSPELVHALEFLELISVNLLL 103
> YML076c
Length=944
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 31/79 (39%), Gaps = 7/79 (8%)
Query 20 INGCCHGELDALYEMLQQLEEKENIKVDLL--ICCGDFQSLRSEADLQYFHAPP-----K 72
I GC + Y + EK K + +CC + +D+ + P K
Sbjct 45 IEGCSKDDPRLSYPTKLETTEKGKTKRNSFACVCCHSLKQKCEPSDVNDIYRKPCRRCLK 104
Query 73 YKKLKDFHLYFRGLKRAPR 91
+KKL F L R KR PR
Sbjct 105 HKKLCKFDLSKRTRKRKPR 123
> HsM4885113
Length=1288
Score = 27.7 bits (60), Expect = 6.6, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 13/21 (61%), Gaps = 0/21 (0%)
Query 6 LGSSGFLPEKMKVAINGCCHG 26
LG G + EK VAI+ CCH
Sbjct 619 LGDLGAIDEKYDVAISSCCHA 639
> Hs21361252
Length=1288
Score = 27.7 bits (60), Expect = 6.6, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 13/21 (61%), Gaps = 0/21 (0%)
Query 6 LGSSGFLPEKMKVAINGCCHG 26
LG G + EK VAI+ CCH
Sbjct 619 LGDLGAIDEKYDVAISSCCHA 639
> CE05549
Length=903
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 30 ALYEMLQQLEEKENI--KVDLLICCGDFQSLRSEADLQYFHAPPKYKKL 76
A+ ++ QL+ ++ KVD +I CGD S + LQ A PK ++
Sbjct 475 AVQNLISQLQLPASVSAKVDKIIACGDKARKPSRSGLQASQARPKVPEI 523
> Hs4503755
Length=532
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 33/125 (26%), Positives = 53/125 (42%), Gaps = 17/125 (13%)
Query 15 KMKV-AINGCCHGELDALYEMLQQLEEKENIKV-DLLICCGDFQS-----LRSEADL--- 64
K KV ++ C + +E++ EEK+ + D ++ C F + L S +
Sbjct 107 KTKVCSVTKCSDSAVSGQWEVVTMHEEKQESAIFDAVMVCTGFLTNPYLPLDSFPGINAF 166
Query 65 --QYFHAPPKYKK---LKDFHLYFRGLKRAPRLTLLVGGNHEAPNVLRQLYYGGWLAPNI 119
QYFH+ +YK KD + G+ + + V +H A V GGW+ I
Sbjct 167 KGQYFHSR-QYKHPDIFKDKRVLVIGMGNSGT-DIAVEASHLAEKVFLSTTGGGWVISRI 224
Query 120 FFLGY 124
F GY
Sbjct 225 FDSGY 229
> YFL008w
Length=1225
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 26/83 (31%), Positives = 35/83 (42%), Gaps = 12/83 (14%)
Query 28 LDALYEMLQQLEEKENIKVDLLICCGDFQSLRSEADLQYF------HAPPKYKKLKDFHL 81
LDA+Y ++L + N V+L G SL E + + F HA P K+ KD
Sbjct 1075 LDAIY---RELTKNPNSNVEL---AGGNASLTIEDEDEPFNAGIKYHATPPLKRFKDMEY 1128
Query 82 YFRGLKRAPRLTLLVGGNHEAPN 104
G K L LL N P+
Sbjct 1129 LSGGEKTVAALALLFAINSYQPS 1151
> Hs7662390
Length=988
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 5 DLGSSGFLPEKMKVAINGCCHGELDALYEMLQQLEEKE 42
D GS+ P + AI C E + + ++ +L EKE
Sbjct 377 DCGSASMKPSQSASAICECTEAEAEIILQLKSELREKE 414
Lambda K H
0.322 0.143 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40