bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3551_orf1
Length=509
Score E
Sequences producing significant alignments: (Bits) Value
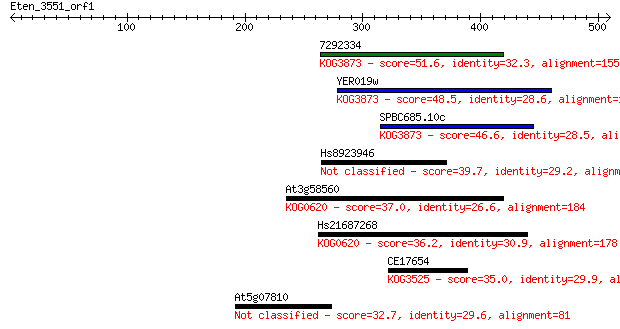
7292334 51.6 4e-06
YER019w 48.5 3e-05
SPBC685.10c 46.6 1e-04
Hs8923946 39.7 0.015
At3g58560 37.0 0.096
Hs21687268 36.2 0.18
CE17654 35.0 0.38
At5g07810 32.7 2.1
> 7292334
Length=402
Score = 51.6 bits (122), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 50/166 (30%), Positives = 77/166 (46%), Gaps = 22/166 (13%)
Query 264 RLHHIPAAILSTGADIICLQEVYGENHADFIIESTRHKLPFVGRRTSGGRFALHNGLMVL 323
R+ I + S DI+ LQEV+ + ++ + + T LP SG + GL+VL
Sbjct 26 RIDAICKELASGKYDIVSLQEVWAQEDSELLQKGTEAVLPHSHYFHSG---VMGAGLLVL 82
Query 324 SKFPIVHTQFHKFEDVTYIERC-----FASKGMLEVAIDVPSIGVIAVFNIHLASGAVDP 378
SK+PI+ T FH + Y R F KG+ I V ++ ++N HL + +
Sbjct 83 SKYPILGTLFHAWSVNGYFHRIQHADWFGGKGVGLCRILVGG-QMVHLYNAHLHAEYDNA 141
Query 379 ESRYVEDVRAAEIRQLLKACDAA----ADRGHVPLVI--GDLNAAP 418
Y + ++++A D A A RG+ L I GDLNA P
Sbjct 142 NDEY-------KTHRVIQAFDTAQFIEATRGNSALQILAGDLNAQP 180
> YER019w
Length=477
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 52/189 (27%), Positives = 82/189 (43%), Gaps = 13/189 (6%)
Query 278 DIICLQEVYGENHADFIIESTRHKLPFVGRRTSGGRFALHNGLMVLSKFPIVHTQFHKFE 337
D+I LQE++ ++ + K P+ +R GL +LSK PI T ++F
Sbjct 94 DVIALQEIWCVEDWKYLASACASKYPY--QRLFHSGILTGPGLAILSKVPIESTFLYRFP 151
Query 338 DVTYIERCFASKGML--EVAIDVPSIGV--IAVFNIHL-ASGAVDPESRYV--EDVRAAE 390
F + +AI V + G IA+ N H+ A A ++ Y+ +A +
Sbjct 152 INGRPSAVFRGDWYVGKSIAITVLNTGTRPIAIMNSHMHAPYAKQGDAAYLCHRSCQAWD 211
Query 391 IRQLLKACDAAADRGHVPLVIGDLNAAPHLCASNYKCFVERGWRDCWLHVHGHQRHLLQH 450
+L+K A G+ +V+GDLN+ P + E G D W +HG Q +
Sbjct 212 FSRLIKLYRQA---GYAVIVVGDLNSRPGSLPHKFLT-QEAGLVDSWEQLHGKQDLAVIA 267
Query 451 HLLHQQQLL 459
L QQLL
Sbjct 268 RLSPLQQLL 276
> SPBC685.10c
Length=368
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 37/139 (26%), Positives = 62/139 (44%), Gaps = 15/139 (10%)
Query 315 ALHNGLMVLSKFPIVHTQFHKFE-----DVTYIERCFASKGMLEVAIDVPSIGVIAVFNI 369
A+ GL + SKFPI+ + +K+ + + KG+ ++ PS +I++FN
Sbjct 21 AMGAGLAMFSKFPIIESSMNKYPLNGRPQAFWRGDWYVGKGVATASLQHPSGRIISLFNT 80
Query 370 HLASGAVDPESRYVEDVRAAEIRQLLKACDAAADRGHVPLVIGDLN----AAPHLCASNY 425
HL + Y+ R ++ + K AA RGH+ + GD N + PH ++Y
Sbjct 81 HLHAPYGKGADTYLCH-RLSQAWYISKLLRAAVQRGHIVIAAGDFNIQPLSVPHEIITSY 139
Query 426 KCFVERGWRDCWLHVHGHQ 444
D WL V+ Q
Sbjct 140 GLV-----NDAWLSVYPDQ 153
> Hs8923946
Length=655
Score = 39.7 bits (91), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 52/124 (41%), Gaps = 23/124 (18%)
Query 265 LHHIPAAILSTGADIICLQEVYGENHADFIIESTRHKLPFV-------GRRTSGGRFALH 317
H +A D +CLQEV+ + A + E ++ G + L+
Sbjct 345 FDHEVSAFFPANLDFLCLQEVFDKRAATKLKEQLHGYFEYILYDVGVYGCQGCCSFKCLN 404
Query 318 NGLMVLSKFPIVHTQFHKFEDVTYIERC----FASKGMLEVAIDVPS-------IGVIAV 366
+GL+ S++PI+ +H Y +C ASKG L + + V S +G IA
Sbjct 405 SGLLFASRYPIMDVAYH-----CYPNKCNDDALASKGALFLKVQVGSTPQDQRIVGYIAC 459
Query 367 FNIH 370
++H
Sbjct 460 THLH 463
> At3g58560
Length=597
Score = 37.0 bits (84), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 49/219 (22%), Positives = 85/219 (38%), Gaps = 45/219 (20%)
Query 235 LSLLSFNAGLLEYKLCGLRVYQNPP-----FTQRRLHHIPAAILSTGADIICLQEVYGEN 289
++LS+N +L +Y P +T RR ++ I+ ADI+CLQEV ++
Sbjct 248 FTVLSYN--ILSDTYASSDIYSYCPTWALAWTYRR-QNLLREIVKYRADIVCLQEVQNDH 304
Query 290 HADFIIESTRHKLPFVGRRTSGGRFALHNGLMVLSKFPIVH--TQFHKFEDVTYIERCFA 347
+F LP + + G F + + + F + + +++E A
Sbjct 305 FEEFF-------LPELDKHGYQGLFKRKTNEVFIGNTNTIDGCATFFRRDRFSHVEFNKA 357
Query 348 SKGMLEVAIDVP------------SIGVIAVFNIHLASGAVD-PESRY------------ 382
++ + E I V ++ +I V S A D P R
Sbjct 358 AQSLTEAIIPVSQKKNALNRLVKDNVALIVVLEAKFGSQAADNPGKRQLLCVANTHVNVP 417
Query 383 --VEDVRAAEIRQLLKACDAAADRGHVP-LVIGDLNAAP 418
++DV+ ++ LLK + A +P LV GD N P
Sbjct 418 HELKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNTVP 456
> Hs21687268
Length=475
Score = 36.2 bits (82), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 55/237 (23%), Positives = 91/237 (38%), Gaps = 67/237 (28%)
Query 262 QRRLHHIPAAILSTGADIICLQEVYGENHADFIIESTRHKLPFVGRRTSGGRFALHNGLM 321
+ R I I++ ADII LQEV E + LP + R G F+ +
Sbjct 191 EYRKKGIMEEIVNCDADIISLQEVETEQYFTLF-------LPALKERGYDGFFSPKSRAK 243
Query 322 VLS-----------------KFPIVHTQFHKFEDVTYIERCFASKGMLEVAIDVPSIGVI 364
++S KF +V +F V + S+ ML + +IGV
Sbjct 244 IMSEQERKHVDGCAIFFKTEKFTLVQKHTVEFNQVA-MANSDGSEAMLNRVMTKDNIGVA 302
Query 365 AVFNIH-------------------LASGA---VDPESRYVEDVR----AAEIRQLLKAC 398
V +H + + A DPE V+ ++ +E++ +L+
Sbjct 303 VVLEVHKELFGAGMKPIHAADKQLLIVANAHMHWDPEYSDVKLIQTMMFVSEVKNILEKA 362
Query 399 DA-----AADRGHVPLVI-GDLNAAP------HL----CASNYKCFVERGWRDCWLH 439
+ AD +PLV+ DLN+ P +L A N+K F E G+ +C ++
Sbjct 363 SSRPGSPTADPNSIPLVLCADLNSLPDSGVVEYLSNGGVADNHKDFKELGYNECLMN 419
> CE17654
Length=760
Score = 35.0 bits (79), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 322 VLSKFPIVHTQFHKFEDVTYIERCFASKGMLEVAIDVPSIG--VIAVFNIHLASGAVDPE 379
+ KFP T+F KF + IE+ F + L+V ++P+ + +++ GA + +
Sbjct 561 IFQKFPGTSTRFQKFPKILEIEKMFRNSKKLQVFRNIPNGNRLQLQLYSDGCYGGADENK 620
Query 380 SRYVEDVRA 388
YVE V+A
Sbjct 621 VSYVEHVQA 629
> At5g07810
Length=1178
Score = 32.7 bits (73), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 40/81 (49%), Gaps = 10/81 (12%)
Query 192 GEDSRSPPDAQSARTDETEAATPSRGASTMQGDDRTTQTRLDELSLLSFNAGLLEYKLCG 251
G+D R P Q+ R +E EA+ PS+G + + + T + L++L F + E+K
Sbjct 828 GKDPRPRPHFQNFRPEEIEASNPSQGTTKEKNPESITDDPVHVLAILEF---MKEWK--S 882
Query 252 LRVYQNPPFTQRRLHHIPAAI 272
LR P +R+L P +
Sbjct 883 LR-----PIEKRKLLGKPLQL 898
Lambda K H
0.318 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13757248296
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40