bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3546_orf2
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
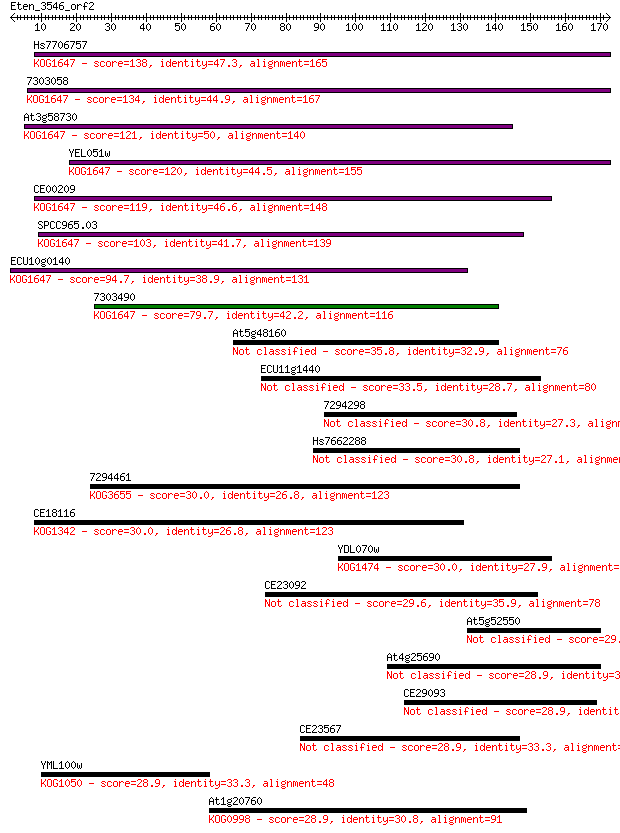
Hs7706757 138 4e-33
7303058 134 7e-32
At3g58730 121 7e-28
YEL051w 120 1e-27
CE00209 119 3e-27
SPCC965.03 103 1e-22
ECU10g0140 94.7 8e-20
7303490 79.7 3e-15
At5g48160 35.8 0.042
ECU11g1440 33.5 0.23
7294298 30.8 1.4
Hs7662288 30.8 1.6
7294461 30.0 2.2
CE18116 30.0 2.3
YDL070w 30.0 2.6
CE23092 29.6 2.7
At5g52550 29.6 3.0
At4g25690 28.9 4.8
CE29093 28.9 4.9
CE23567 28.9 5.0
YML100w 28.9 5.3
At1g20760 28.9 5.6
> Hs7706757
Length=247
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 78/168 (46%), Positives = 114/168 (67%), Gaps = 8/168 (4%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V + A DNVAGV LPVF+ + T D + L+ GG + + Y +A+
Sbjct 84 IQNVNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLARGGEQLAKLKRNYAKAV 142
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++++++ YI ELDE EREEF+RL
Sbjct 143 ELLVELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLAYIITELDEREREEFYRL 202
Query 128 KKIQEKKRLAKEAEQKALEE---AGKALKASGLLSGTILGEDDDDLVF 172
KKIQEKK++ KE +K LE+ AG+ L+ + LL+ E D+DL+F
Sbjct 203 KKIQEKKKILKEKSEKDLEQRRAAGEVLEPANLLA----EEKDEDLLF 246
> 7303058
Length=246
Score = 134 bits (338), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 75/170 (44%), Positives = 111/170 (65%), Gaps = 8/170 (4%)
Query 6 LFLQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQE 65
+ LQ + + + + DNVAGV LPVF+ D + D + L+ GG + + YQ
Sbjct 82 VVLQNVTKAQIKIRTKKDNVAGVTLPVFESYQDGS-DTYELAGLARGGQQLAKLKKNYQS 140
Query 66 ALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFF 125
A+ LV+LASLQT+F TLD IK+TNRRVNA+ +V++P++D+++ YI ELDE+EREEF+
Sbjct 141 AVKLLVELASLQTSFVTLDEVIKITNRRVNAIEHVIIPRIDRTLAYIISELDELEREEFY 200
Query 126 RLKKIQEKKRLAK---EAEQKALEEAGKALKASGLLSGTILGEDDDDLVF 172
RLKKIQ+KKR A+ +A++ L + G ++ IL E DDD++F
Sbjct 201 RLKKIQDKKREARIKADAKKAELLQQGIDVRQ----QANILDEGDDDVLF 246
> At3g58730
Length=261
Score = 121 bits (303), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 70/141 (49%), Positives = 94/141 (66%), Gaps = 5/141 (3%)
Query 5 WLFLQRIKRPAVFVTAAYDNVAGVRLPVF-QITTDPTVDILKNINLSAGGHVILAARDKY 63
+ L+ +K + V + +N+AGV+LP F + T + L L+ GG + A R Y
Sbjct 83 HVVLENVKEATLKVRSRTENIAGVKLPKFDHFSEGETKNDL--TGLARGGQQVRACRVAY 140
Query 64 QEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREE 123
+A+ LV+LASLQT+F TLD IK TNRRVNAL NVV PKL+ +I+YI ELDE+ERE+
Sbjct 141 VKAIEVLVELASLQTSFLTLDEAIKTTNRRVNALENVVKPKLENTISYIKGELDELERED 200
Query 124 FFRLKKIQEKKRLAKEAEQKA 144
FFRLKKIQ KR +E E++A
Sbjct 201 FFRLKKIQGYKR--REVERQA 219
> YEL051w
Length=256
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 69/163 (42%), Positives = 103/163 (63%), Gaps = 8/163 (4%)
Query 18 VTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQ 77
V A +NV+GV L F+ DP ++ + L GG + A++ Y A+ LV+LASLQ
Sbjct 94 VRARQENVSGVYLSQFESYIDPEINDFRLTGLGRGGQQVQRAKEIYSRAVETLVELASLQ 153
Query 78 TAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKR-- 135
TAF LD IK+TNRRVNA+ +V++P+ + +I YI ELDE++REEF+RLKK+QEKK+
Sbjct 154 TAFIILDEVIKVTNRRVNAIEHVIIPRTENTIAYINSELDELDREEFYRLKKVQEKKQNE 213
Query 136 LAK-EAEQK-----ALEEAGKALKASGLLSGTILGEDDDDLVF 172
AK +AE K A ++A + T++ + +DD++F
Sbjct 214 TAKLDAEMKLKRDRAEQDASEVAADEEPQGETLVADQEDDVIF 256
> CE00209
Length=257
Score = 119 bits (298), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 69/150 (46%), Positives = 91/150 (60%), Gaps = 3/150 (2%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEAL 67
+Q + + V +NV GV LPVF D D L GG I + Y +A+
Sbjct 86 IQNVSQAQYRVRMKKENVVGVFLPVFDAYQDGP-DAYDLTGLGKGGANIARLKKNYNKAI 144
Query 68 AELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRL 127
LV+LA+LQT F TLD IK+TNRRVNA+ +V++P+++ ++ YI ELDEMEREEFFR+
Sbjct 145 ELLVELATLQTCFITLDEAIKVTNRRVNAIEHVIIPRIENTLTYIVTELDEMEREEFFRM 204
Query 128 KKIQEKKRLAKEAE--QKALEEAGKALKAS 155
KKIQ K+ KE E QKALE G A+
Sbjct 205 KKIQANKKKLKEQEAAQKALEGPGPGEDAA 234
> SPCC965.03
Length=285
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 58/139 (41%), Positives = 94/139 (67%), Gaps = 0/139 (0%)
Query 9 QRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALA 68
Q +K+P + V + +N++GV LP F++ D ++D + L GG I AR Y++A+
Sbjct 86 QSVKQPRLRVRSKQENISGVFLPTFEMNLDESIDDFQLTGLGKGGQQIQKARQVYEKAVE 145
Query 69 ELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLK 128
LV+LAS Q+AF L ++MTNRRVN++ ++++P+L+ +I YI EL+E+ERE+F RLK
Sbjct 146 TLVQLASYQSAFVLLGDVLQMTNRRVNSIEHIIIPRLENTIKYIESELEELEREDFTRLK 205
Query 129 KIQEKKRLAKEAEQKALEE 147
K+Q+ K A++A+ EE
Sbjct 206 KVQKTKENAEKADSVTKEE 224
> ECU10g0140
Length=212
Score = 94.7 bits (234), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 51/131 (38%), Positives = 78/131 (59%), Gaps = 3/131 (2%)
Query 1 GLGLWLFLQRIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAAR 60
G L +FL ++ V+V + + V+GV LP F + + IL L G + R
Sbjct 75 GANLKMFLYECQKQNVYVRSRVEQVSGVSLPFFSLQKENIQPIL---FLDRSGQSLNECR 131
Query 61 DKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEME 120
+K+ E L LV L +L+ +F L+S + TNRRVNAL ++P+L+ +++YI ELDE +
Sbjct 132 EKFLEVLEMLVDLCALKNSFRVLNSILMSTNRRVNALEFNIIPRLENTVSYIVSELDEQD 191
Query 121 REEFFRLKKIQ 131
R +FFRLKK+Q
Sbjct 192 RGDFFRLKKVQ 202
> 7303490
Length=373
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 49/116 (42%), Positives = 67/116 (57%), Gaps = 1/116 (0%)
Query 25 VAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQTAFFTLD 84
+ GV+L ++ T V LS GG + RD Y +AL LV+ ASL+ L+
Sbjct 101 IVGVKLNTLELETK-GVGAFPLAGLSCGGMQVSRIRDSYTKALKALVEFASLEYQVRMLE 159
Query 85 SEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKEA 140
+ TN RVNAL +VV+P L + NYI EL+E ERE+F+RLK+ Q K+ AK A
Sbjct 160 AASLQTNMRVNALEHVVIPILQNTYNYICGELEEFEREDFYRLKRSQAKQLEAKMA 215
> At5g48160
Length=574
Score = 35.8 bits (81), Expect = 0.042, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 41/83 (49%), Gaps = 9/83 (10%)
Query 65 EALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKS-----INYITKELDEM 119
+ L +V+L + F L + R + L +VL K+DKS NY+ + L E
Sbjct 451 DELERIVRLKQAEADMFQLKA--NEAKREADRLQRIVLAKMDKSEEEYASNYLKQRLSEA 508
Query 120 EREEFFRLKKI--QEKKRLAKEA 140
E E+ + +KI QE R+A ++
Sbjct 509 EAEKQYLFEKIKLQENSRVASQS 531
> ECU11g1440
Length=1303
Score = 33.5 bits (75), Expect = 0.23, Method: Composition-based stats.
Identities = 23/87 (26%), Positives = 43/87 (49%), Gaps = 7/87 (8%)
Query 73 LASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQE 132
LA + +++K R+ NALN + +K+I + + +EM++E K+ E
Sbjct 517 LAKAHENLENVKTQLKPGGRKTNALNFLSKVLTEKAIKDVNQRNEEMQKEAVKAQKEANE 576
Query 133 -------KKRLAKEAEQKALEEAGKAL 152
++ LAK AEQ+ + GK++
Sbjct 577 NSAVKSAEEHLAKAAEQQIINNEGKSV 603
> 7294298
Length=535
Score = 30.8 bits (68), Expect = 1.4, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 34/55 (61%), Gaps = 4/55 (7%)
Query 91 NRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKEAEQKAL 145
N+ ++ L+ K+D+ T+ LD +++F RLK+ ++++R+ ++ +Q AL
Sbjct 427 NQSIDTLDE----KVDEENEGDTRTLDSRNKDDFDRLKEKRDRERVNRQMQQAAL 477
> Hs7662288
Length=967
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query 88 KMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQ-EKKRLAKEAEQKALE 146
++ + ++++ L LD + TKEL+E++ EE +RL+++ LA + E+ L+
Sbjct 627 ELKAKEIDSMQKQRLDWLDAETSRRTKELNELKAEEMYRLQQLSVSATHLADKVEEAVLD 686
> 7294461
Length=531
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 62/127 (48%), Gaps = 15/127 (11%)
Query 24 NVAGVRLPVFQITTDPTVDILKNI-NLSAGGHVILAARDKYQEALAELVK-LASLQTAFF 81
N G PV + T + ++++ NL +G H+ + AR++ L L+K L+++ +A+
Sbjct 83 NWQGEGAPVLRKGT--CANHIRDVSNLLSGAHLTINARNEDDIDLDRLLKKLSTVSSAYS 140
Query 82 TLDSEIKMTNRRVNALNNV--VLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKE 139
+ M ++ N V+P TKEL+ ++F++ ++ +EK R E
Sbjct 141 FKEPRGAMEEQKAPVGTNYTRVIP---------TKELNASVMQDFWKKEEAEEKLRQEAE 191
Query 140 AEQKALE 146
E K LE
Sbjct 192 KESKRLE 198
> CE18116
Length=465
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 33/137 (24%), Positives = 63/137 (45%), Gaps = 14/137 (10%)
Query 8 LQRIKRPAVFVTAAYDNVAGVRLPVFQITT---DPTVDILKNIN-----LSAGGHVILAA 59
+ R + AV + D++AG RL VF +TT V+ +K+ N + GG+ I
Sbjct 248 MARFQPEAVVLQCGADSLAGDRLGVFNLTTYGHGKCVEYMKSFNVPLLLVGGGGYTIRNV 307
Query 60 RDK--YQEALA---ELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPK-LDKSINYIT 113
Y+ A+A E+ L F + K+ + + AL+N P+ +D++I +
Sbjct 308 SRCWLYETAIALNQEVSDDLPLHDYFDYFIPDYKLHIKPLAALSNFNTPEFIDQTIVALL 367
Query 114 KELDEMEREEFFRLKKI 130
+ L ++ +++ I
Sbjct 368 ENLKQLPHVPSVQMQSI 384
> YDL070w
Length=638
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 95 NALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQEKKRLAKE-AEQKALEEAGKALK 153
N + N + L++ + + EL +++R+E +L K +++K L K +KA++ + LK
Sbjct 462 NDITNPAIQYLEQKLKKMEVELQQLKRQELSKLSKERKRKHLGKTLLRRKAMKHSVDDLK 521
Query 154 AS 155
S
Sbjct 522 KS 523
> CE23092
Length=2129
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 42/100 (42%), Gaps = 22/100 (22%)
Query 74 ASLQTAFFTLDSEIKMTNRRVNALNNVVLPKLDKSINYITKELD---EMEREEFFRLKKI 130
A ++ F L ++ + + + +L ++ +Y K D E ER++F L KI
Sbjct 996 AQVRELFEHLANQTADKGKELRKFLSSILKEIHPPEDYCIKVYDPIGEEERKQFAILAKI 1055
Query 131 Q-------------------EKKRLAKEAEQKALEEAGKA 151
Q EK RL KEA Q+ LEE KA
Sbjct 1056 QAEEKQRRLDEAKEKLAIDKEKLRLKKEALQRELEERNKA 1095
> At5g52550
Length=197
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 24/38 (63%), Gaps = 1/38 (2%)
Query 132 EKKRLAKEAEQKALEEAGKALKASGLLSGTILGEDDDD 169
EKK+L K EQ+ L+E G A+ A + +LGED DD
Sbjct 55 EKKKLRKLEEQRRLDEEGAAI-AEAVALHVLLGEDCDD 91
> At4g25690
Length=191
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 41/81 (50%), Gaps = 21/81 (25%)
Query 109 INYITKELDEMEREEFFRLKKIQEKKRLAKEA--------------------EQKALEEA 148
+ +T+++ + ++EF R+K+ ++KKR ++A EQ+ L+E
Sbjct 10 VQPVTRKVKKHGKDEFDRIKQAEKKKRRLEKALATSAAIRAELEKKKQKRLEEQQRLDEE 69
Query 149 GKALKASGLLSGTILGEDDDD 169
G A+ A + +LGED DD
Sbjct 70 GAAI-AEAVALHVLLGEDSDD 89
> CE29093
Length=772
Score = 28.9 bits (63), Expect = 4.9, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 12/67 (17%)
Query 114 KELDEMEREEFFRLKKIQEKKRLAKEAEQKALEEAGK-----------ALKASGL-LSGT 161
K+ +E R + KK +++K AK EQ+ +EEA K L+A L L T
Sbjct 483 KKQEEEIRAKLEAKKKAEQEKERAKREEQRRIEEAAKLEYRRRIEESQRLEAERLALEAT 542
Query 162 ILGEDDD 168
++ ED+D
Sbjct 543 MVDEDED 549
> CE23567
Length=1014
Score = 28.9 bits (63), Expect = 5.0, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 10/65 (15%)
Query 84 DSEIKMTNRRVNALNNVVLPKLDKSINYITKELDEMEREEFFRLKKIQE--KKRLAKEAE 141
+SE++ ++ VN +NN ++ L K IN KEL E F L + E K+ ++ + E
Sbjct 591 NSEVQNKDKEVNEINNKIIQHLKKIIN--AKELGE------FTLPMLNECIKEVISNDNE 642
Query 142 QKALE 146
K +E
Sbjct 643 NKTIE 647
> YML100w
Length=1098
Score = 28.9 bits (63), Expect = 5.3, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 3/48 (6%)
Query 10 RIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVIL 57
RI P + +D+ + + P+ Q DPT ++LKN+N S H +L
Sbjct 144 RIASP---IQHEHDSGSRIASPIQQQQQDPTTNLLKNVNKSLLVHSLL 188
> At1g20760
Length=1019
Score = 28.9 bits (63), Expect = 5.6, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 6/94 (6%)
Query 58 AARDKYQEALAELVKLASLQTAFFTLDSEIKMTNRRVNALNNVVLPK--LDKSINYITKE 115
+A EA A+ K+ Q A+ +DS K+ R + ++VL K D +N I++
Sbjct 526 SASSNLPEAAADEEKVDEKQNAY--MDSREKLDYYRTK-MQDIVLYKSRCDNRLNEISER 582
Query 116 LDEMEREEFFRLKKIQEK-KRLAKEAEQKALEEA 148
+RE KK +EK K++A+ + +EEA
Sbjct 583 ASADKREAETLAKKYEEKYKQVAEIGSKLTIEEA 616
Lambda K H
0.318 0.136 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40