bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3533_orf1
Length=58
Score E
Sequences producing significant alignments: (Bits) Value
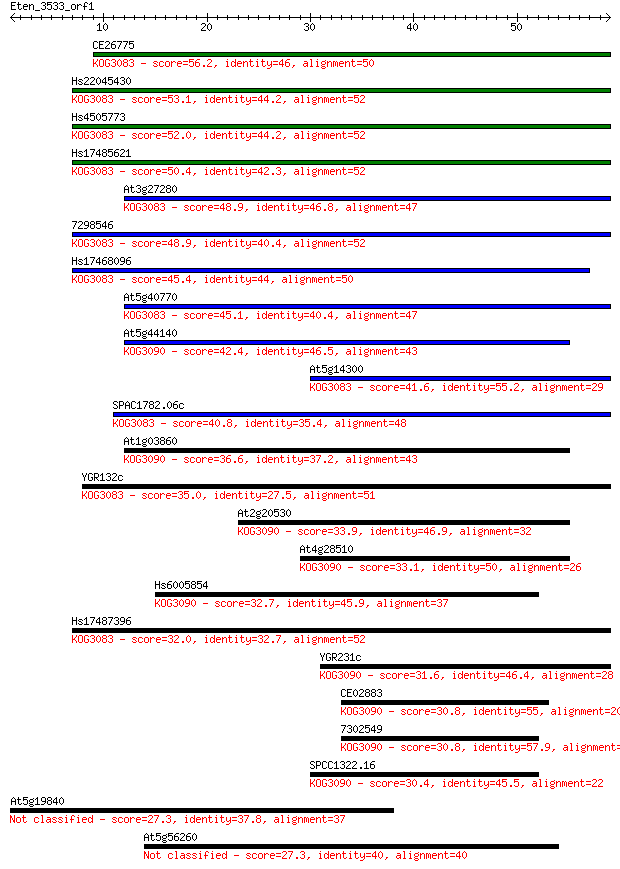
CE26775 56.2 2e-08
Hs22045430 53.1 1e-07
Hs4505773 52.0 2e-07
Hs17485621 50.4 8e-07
At3g27280 48.9 2e-06
7298546 48.9 2e-06
Hs17468096 45.4 3e-05
At5g40770 45.1 3e-05
At5g44140 42.4 2e-04
At5g14300 41.6 3e-04
SPAC1782.06c 40.8 6e-04
At1g03860 36.6 0.012
YGR132c 35.0 0.035
At2g20530 33.9 0.079
At4g28510 33.1 0.14
Hs6005854 32.7 0.17
Hs17487396 32.0 0.27
YGR231c 31.6 0.38
CE02883 30.8 0.58
7302549 30.8 0.58
SPCC1322.16 30.4 0.89
At5g19840 27.3 6.2
At5g56260 27.3 7.8
> CE26775
Length=275
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 23/50 (46%), Positives = 37/50 (74%), Gaps = 0/50 (0%)
Query 9 ERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
+++L LG +GV + ++G +A++ LY+VDGGQR ++F+RF GV VGE
Sbjct 6 QKLLGRLGTVGVGLSIAGGIAQTALYNVDGGQRAVIFDRFSGVKNEVVGE 55
> Hs22045430
Length=236
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 23/52 (44%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 7 MTERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
M +V ++G+ G+A+ V+G V S LY+VD G R ++F+RF GV VGE
Sbjct 1 MAAKVFASIGKFGLALAVAGGVVNSALYNVDAGHRAVIFDRFHGVQDIVVGE 52
> Hs4505773
Length=272
Score = 52.0 bits (123), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/52 (44%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 7 MTERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
M +V ++G+ G+A+ V+G V S LY+VD G R ++F+RF GV VGE
Sbjct 1 MAAKVFESIGKFGLALAVAGGVVNSALYNVDAGHRAVIFDRFRGVQDIVVGE 52
> Hs17485621
Length=250
Score = 50.4 bits (119), Expect = 8e-07, Method: Composition-based stats.
Identities = 22/52 (42%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 7 MTERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
M +V ++G+ G+A+ V G + S LY+VD G R ++F+RF GV VGE
Sbjct 1 MAAKVFVSIGKFGLALAVVGSMLNSALYNVDAGHRAVIFDRFCGVQDIVVGE 52
> At3g27280
Length=279
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/47 (46%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 12 LTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
LTNL + +GV+ S LY VDGG+R ++F+RF GV + VGE
Sbjct 11 LTNLAKAAFGLGVAATALNSSLYTVDGGERAVLFDRFRGVLDQTVGE 57
> 7298546
Length=276
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 32/52 (61%), Gaps = 0/52 (0%)
Query 7 MTERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
M + +G++G+ V V G V S LY+V+GG R ++F+RF G+ VGE
Sbjct 1 MAAQFFNRIGQMGLGVAVLGGVVNSALYNVEGGHRAVIFDRFTGIKENVVGE 52
> Hs17468096
Length=249
Score = 45.4 bits (106), Expect = 3e-05, Method: Composition-based stats.
Identities = 22/53 (41%), Positives = 32/53 (60%), Gaps = 3/53 (5%)
Query 7 MTERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVS---PRPV 56
M ++ +G+ G+A+ V+G V S LY VD G R ++F+RF GV PR V
Sbjct 1 MAAKMFEFIGKFGLALVVAGGVVNSALYSVDAGHRAVVFDRFRGVQDIVPRNV 53
> At5g40770
Length=277
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 12 LTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
L+NL + +G + V + L+ VDGG+R ++F+RF GV + VGE
Sbjct 11 LSNLAKAAFGLGTAATVLNTSLFTVDGGERAVIFDRFRGVMDQTVGE 57
> At5g44140
Length=278
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 20/43 (46%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 12 LTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPR 54
L+ L +LGV G+ S +Y+VDGG R I+FNRF G+ R
Sbjct 15 LSALLKLGVIGGLGLYCIGSSMYNVDGGHRAIVFNRFTGIKDR 57
> At5g14300
Length=249
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 16/29 (55%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 30 KSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
+S ++ VDGGQR +MF+RF G+ PVGE
Sbjct 21 RSTMFTVDGGQRAVMFHRFEGILEEPVGE 49
> SPAC1782.06c
Length=282
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 11 VLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
VL + R + +G+ + +S +YDV GG+R ++F+R GV + V E
Sbjct 4 VLERVARYAIPIGIGFTLLQSSIYDVPGGKRAVLFDRLSGVQKQVVQE 51
> At1g03860
Length=286
Score = 36.6 bits (83), Expect = 0.012, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 12 LTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPR 54
L+ L ++ V G+ + LY+VDGG R +MFNR G+ +
Sbjct 15 LSALLKVSVIGGLGVYALTNSLYNVDGGHRAVMFNRLTGIKEK 57
> YGR132c
Length=287
Score = 35.0 bits (79), Expect = 0.035, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 8 TERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
+ +++ + ++ + +G+ + +YDV GG R ++F+R GV + VGE
Sbjct 4 SAKLIDVITKVALPIGIIASGIQYSMYDVKGGSRGVIFDRINGVKQQVVGE 54
> At2g20530
Length=286
Score = 33.9 bits (76), Expect = 0.079, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 23 GVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPR 54
G+S A LY+VDGG R I+FNR G+ +
Sbjct 24 GLSLYGATHTLYNVDGGHRAIVFNRLVGIKDK 55
> At4g28510
Length=288
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 29 AKSCLYDVDGGQRCIMFNRFGGVSPR 54
A LY+V+GG R IMFNR G+ +
Sbjct 32 ATHSLYNVEGGHRAIMFNRLVGIKDK 57
> Hs6005854
Length=299
Score = 32.7 bits (73), Expect = 0.17, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 21/37 (56%), Gaps = 5/37 (13%)
Query 15 LGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGV 51
LG VA GV + ++ V+GG R I FNR GGV
Sbjct 27 LGAGAVAYGV-----RESVFTVEGGHRAIFFNRIGGV 58
> Hs17487396
Length=376
Score = 32.0 bits (71), Expect = 0.27, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 6/52 (11%)
Query 7 MTERVLTNLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
M +V + + G+A+ V+G + +VD G R ++F+ F GV VGE
Sbjct 1 MAAKVFEFISKFGLALAVAGGL------NVDAGHRAVIFDLFRGVQDIVVGE 46
> YGR231c
Length=315
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 31 SCLYDVDGGQRCIMFNRFGGVSPRPVGE 58
+ L++VDGG R I+++R GVS R E
Sbjct 56 NALFNVDGGHRAIVYSRIHGVSSRIFNE 83
> CE02883
Length=286
Score = 30.8 bits (68), Expect = 0.58, Method: Composition-based stats.
Identities = 11/20 (55%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 33 LYDVDGGQRCIMFNRFGGVS 52
++ V+ G R IMFNR GG+S
Sbjct 32 MFTVEAGHRAIMFNRIGGLS 51
> 7302549
Length=303
Score = 30.8 bits (68), Expect = 0.58, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 15/19 (78%), Gaps = 0/19 (0%)
Query 33 LYDVDGGQRCIMFNRFGGV 51
LY V+GG R I+F+R GG+
Sbjct 42 LYTVEGGHRAIIFSRLGGI 60
> SPCC1322.16
Length=279
Score = 30.4 bits (67), Expect = 0.89, Method: Composition-based stats.
Identities = 10/22 (45%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 30 KSCLYDVDGGQRCIMFNRFGGV 51
++ L++VDGG R I ++R GG+
Sbjct 33 QTSLFNVDGGHRAIKYSRIGGI 54
> At5g19840
Length=399
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 1 DRSFLEMTERVLTNLGRLGVAVGVSGLVAKSCLYDVD 37
D+S +EM E N+G + G+S L K+ L+D+D
Sbjct 233 DQSRIEMAEGGNDNIGNESIKKGLSTLHEKASLHDLD 269
> At5g56260
Length=166
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 14 NLGRLGVAVGVSGLVAKSCLYDVDGGQRCIMFNRFGGVSP 53
NLG+L G SG+V C+ DVD C + R G +P
Sbjct 83 NLGQLAQNNGWSGIVVNGCVRDVDEINDCDVGVRALGSNP 122
Lambda K H
0.324 0.144 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187999082
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40