bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3518_orf1
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
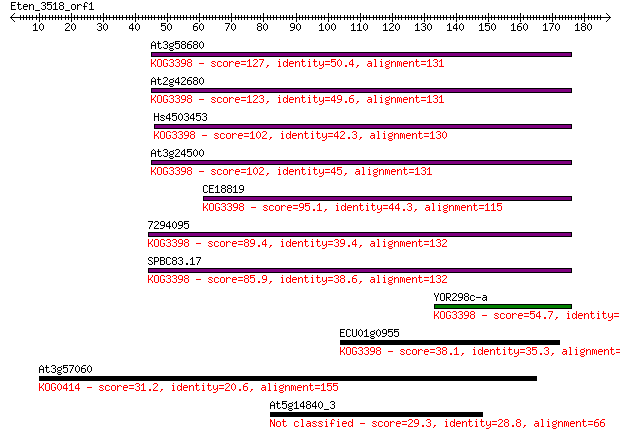
At3g58680 127 2e-29
At2g42680 123 2e-28
Hs4503453 102 4e-22
At3g24500 102 6e-22
CE18819 95.1 7e-20
7294095 89.4 4e-18
SPBC83.17 85.9 5e-17
YOR298c-a 54.7 1e-07
ECU01g0955 38.1 0.009
At3g57060 31.2 1.4
At5g14840_3 29.3 4.4
> At3g58680
Length=142
Score = 127 bits (318), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 66/131 (50%), Positives = 86/131 (65%), Gaps = 1/131 (0%)
Query 45 QDWTPVVLQKTSPGGGKRVSKEAEVNKARRAGEVIETEKKFLGGRNKTTKAGLIPNAKKV 104
QDW PVV++K +P + E VN ARR+G IET +KF G NK +G N KK+
Sbjct 9 QDWEPVVIRKRAPNAAAK-RDEKTVNAARRSGADIETVRKFNAGSNKAASSGTSLNTKKL 67
Query 105 EEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVIL 164
++DT + DRV T+ +A+ +AR K +TQ+QLA INEKP V+ EYESGKAIPN IL
Sbjct 68 DDDTENLSHDRVPTELKKAIMQARGEKKLTQSQLAHLINEKPQVIQEYESGKAIPNQQIL 127
Query 165 QKMSRALGCQL 175
K+ RALG +L
Sbjct 128 SKLERALGAKL 138
> At2g42680
Length=142
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 65/131 (49%), Positives = 85/131 (64%), Gaps = 1/131 (0%)
Query 45 QDWTPVVLQKTSPGGGKRVSKEAEVNKARRAGEVIETEKKFLGGRNKTTKAGLIPNAKKV 104
QDW PVV++K P E VN ARR+G IET +KF G NK +G N K +
Sbjct 9 QDWEPVVIRK-KPANAAAKRDEKTVNAARRSGADIETVRKFNAGTNKAASSGTSLNTKML 67
Query 105 EEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVIL 164
++DT + +RV T+ +A+ +AR +K +TQ+QLAQ INEKP V+ EYESGKAIPN IL
Sbjct 68 DDDTENLTHERVPTELKKAIMQARTDKKLTQSQLAQIINEKPQVIQEYESGKAIPNQQIL 127
Query 165 QKMSRALGCQL 175
K+ RALG +L
Sbjct 128 SKLERALGAKL 138
> Hs4503453
Length=148
Score = 102 bits (255), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 55/131 (41%), Positives = 85/131 (64%), Gaps = 4/131 (3%)
Query 46 DW-TPVVLQKTSPGGGKRVSKEAEVNKARRAGEVIETEKKFLGGRNKTTKAGLIPNAKKV 104
DW T VL+K P + SK+A + A+R GE +ET KK+ G+NK + + N K+
Sbjct 5 DWDTVTVLRKKGPTAAQAKSKQA-ILAAQRRGEDVETSKKWAAGQNK--QHSITKNTAKL 61
Query 105 EEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVIL 164
+ +T + H DRV+ + + + + R++KG+TQ LA INEKP V+++YESG+AIPN +L
Sbjct 62 DRETEELHHDRVTLEVGKVIQQGRQSKGLTQKDLATKINEKPQVIADYESGRAIPNNQVL 121
Query 165 QKMSRALGCQL 175
K+ RA+G +L
Sbjct 122 GKIERAIGLKL 132
> At3g24500
Length=148
Score = 102 bits (253), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 59/133 (44%), Positives = 79/133 (59%), Gaps = 3/133 (2%)
Query 45 QDWTPVVLQKTSPGGGKRVSKEAEVNKARRAGEVIETEKKFLGGRNKTTKAGLIP--NAK 102
QDW PVVL K S + + VN A R G ++T KKF G NK K+ +P N K
Sbjct 11 QDWEPVVLHK-SKQKSQDLRDPKAVNAALRNGVAVQTVKKFDAGSNKKGKSTAVPVINTK 69
Query 103 KVEEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGV 162
K+EE+T +DRV + + +AR K M+QA LA+ INE+ VV EYE+GKA+PN
Sbjct 70 KLEEETEPAAMDRVKAEVRLMIQKARLEKKMSQADLAKQINERTQVVQEYENGKAVPNQA 129
Query 163 ILQKMSRALGCQL 175
+L KM + LG +L
Sbjct 130 VLAKMEKVLGVKL 142
> CE18819
Length=156
Score = 95.1 bits (235), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 51/115 (44%), Positives = 71/115 (61%), Gaps = 2/115 (1%)
Query 61 KRVSKEAEVNKARRAGEVIETEKKFLGGRNKTTKAGLIPNAKKVEEDTGDYHIDRVSTDF 120
K + A++N A+RAG I TEKK + G N+ A N +++E+T + H +V+
Sbjct 27 KTLKSAAQLNAAQRAGVDISTEKKTMSGGNRQHSAN--KNTLRLDEETEELHHQKVALSL 84
Query 121 CRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGCQL 175
+ + +AR KG TQ L+ INEKP VV EYESGKA+PN I+ KM RALG +L
Sbjct 85 GKVMQQARATKGWTQKDLSTQINEKPQVVGEYESGKAVPNQQIMAKMERALGVKL 139
> 7294095
Length=145
Score = 89.4 bits (220), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 52/136 (38%), Positives = 81/136 (59%), Gaps = 10/136 (7%)
Query 44 FQDWTPV-VLQKTSPGGGKRVSKEAEVNKARRAGEVIETEKKFLGGRNK---TTKAGLIP 99
DW V VL+K +P + E+ VN+ARR G ++T++K+ G NK TTK
Sbjct 1 MSDWDSVTVLRKKAPKSST-LKTESAVNQARRQGVAVDTQQKYGAGTNKQHVTTK----- 54
Query 100 NAKKVEEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIP 159
N K++ +T + D++ D + + + R++KG++Q LA I EK VV++YE+G+ IP
Sbjct 55 NTAKLDRETEELRHDKIPLDVGKLIQQGRQSKGLSQKDLATKICEKQQVVTDYEAGRGIP 114
Query 160 NGVILQKMSRALGCQL 175
N +IL KM R LG +L
Sbjct 115 NNLILGKMERVLGIKL 130
> SPBC83.17
Length=148
Score = 85.9 bits (211), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 51/135 (37%), Positives = 75/135 (55%), Gaps = 5/135 (3%)
Query 44 FQDWTPV--VLQKTSPGGGKRVSK-EAEVNKARRAGEVIETEKKFLGGRNKTTKAGLIPN 100
DW V + + PG V+K ++++N ARRAG ++ TEKK+ G AG +
Sbjct 1 MSDWDTVTKIGSRAGPGARTHVAKTQSQINSARRAGAIVGTEKKYATGNKSQDPAGQ--H 58
Query 101 AKKVEEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPN 160
K++ + +A+ + R+ KG Q L+Q INEKP VV++YESG+AIPN
Sbjct 59 LTKIDRENEVKPPSTTGRSVAQAIQKGRQAKGWAQKDLSQRINEKPQVVNDYESGRAIPN 118
Query 161 GVILQKMSRALGCQL 175
+L KM RALG +L
Sbjct 119 QQVLSKMERALGIKL 133
> YOR298c-a
Length=58
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 33/43 (76%), Gaps = 0/43 (0%)
Query 133 MTQAQLAQAINEKPSVVSEYESGKAIPNGVILQKMSRALGCQL 175
M+Q LA INEKP+VV++YE+ +AIPN +L K+ RALG +L
Sbjct 1 MSQKDLATKINEKPTVVNDYEAARAIPNQQVLSKLERALGVKL 43
> ECU01g0955
Length=95
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 37/68 (54%), Gaps = 3/68 (4%)
Query 104 VEEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVI 163
V EDTG + VS A+A AR KGM++ LAQ + + S++ +E G+A+ N I
Sbjct 24 VPEDTG---LRIVSKRVGDAIANARAQKGMSRKDLAQKMKKNVSIIDSWERGEAVYNEKI 80
Query 164 LQKMSRAL 171
++ L
Sbjct 81 AKEFESIL 88
> At3g57060
Length=1439
Score = 31.2 bits (69), Expect = 1.4, Method: Composition-based stats.
Identities = 32/155 (20%), Positives = 55/155 (35%), Gaps = 7/155 (4%)
Query 10 PRTCAQIPAFRFKVPASWFPASPCSAASVEKMSQFQDWTPVVLQKTSPGGGKRVSKEAEV 69
P TC F + P + + K ++ ++ +L T G K +
Sbjct 1138 PYTC-------FGFNKKYIPLTSTPKLADGKQWEYISYSLSLLTFTEKGIKKLIESFKSY 1190
Query 70 NKARRAGEVIETEKKFLGGRNKTTKAGLIPNAKKVEEDTGDYHIDRVSTDFCRALAEARR 129
A V E + + K K L ++ EE +H+++ + AE R
Sbjct 1191 EHALAEDLVTENFRSIINKGKKFAKPELKACIEEFEEKINKFHMEKKEQEETARNAEVHR 1250
Query 130 NKGMTQAQLAQAINEKPSVVSEYESGKAIPNGVIL 164
K T LA K V EY+ G+ + + I+
Sbjct 1251 EKTKTMESLAVLSKVKEEPVEEYDEGEGVSDSEIV 1285
> At5g14840_3
Length=142
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Query 82 EKKFLGGRNKTTKAGLIPNAKKVEEDTGDYHIDRVSTDFCRALAEARRNKGMTQAQLAQA 141
E+ + N T L PN + E+ +G+ H++ + D + A + +T A A
Sbjct 67 EEHPITSLNIPTSLSLAPNQDQEEKSSGERHLENLDDDTTSSAA-----RTITSAIPQTA 121
Query 142 INEKPS 147
INE PS
Sbjct 122 INEIPS 127
Lambda K H
0.315 0.128 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3058524910
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40