bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3498_orf1
Length=76
Score E
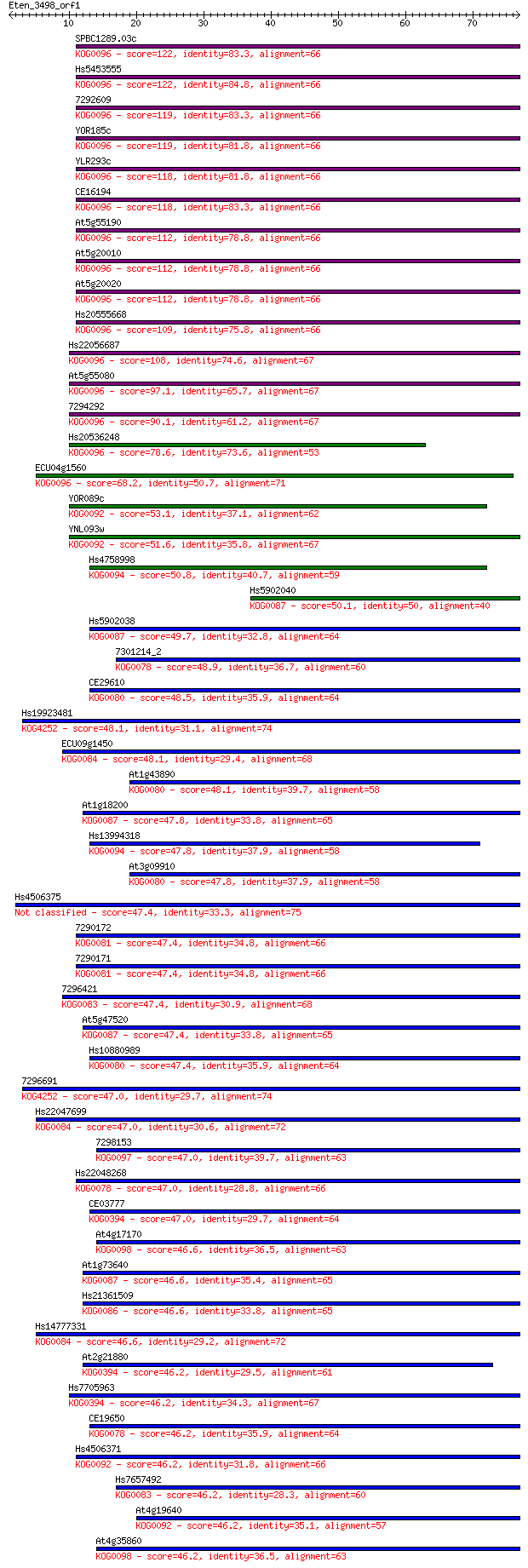
Sequences producing significant alignments: (Bits) Value
SPBC1289.03c 122 2e-28
Hs5453555 122 2e-28
7292609 119 2e-27
YOR185c 119 2e-27
YLR293c 118 2e-27
CE16194 118 2e-27
At5g55190 112 1e-25
At5g20010 112 2e-25
At5g20020 112 2e-25
Hs20555668 109 1e-24
Hs22056687 108 2e-24
At5g55080 97.1 7e-21
7294292 90.1 9e-19
Hs20536248 78.6 3e-15
ECU04g1560 68.2 4e-12
YOR089c 53.1 1e-07
YNL093w 51.6 3e-07
Hs4758998 50.8 7e-07
Hs5902040 50.1 1e-06
Hs5902038 49.7 1e-06
7301214_2 48.9 2e-06
CE29610 48.5 3e-06
Hs19923481 48.1 3e-06
ECU09g1450 48.1 4e-06
At1g43890 48.1 4e-06
At1g18200 47.8 5e-06
Hs13994318 47.8 5e-06
At3g09910 47.8 5e-06
Hs4506375 47.4 6e-06
7290172 47.4 7e-06
7290171 47.4 7e-06
7296421 47.4 7e-06
At5g47520 47.4 7e-06
Hs10880989 47.4 7e-06
7296691 47.0 8e-06
Hs22047699 47.0 8e-06
7298153 47.0 8e-06
Hs22048268 47.0 8e-06
CE03777 47.0 1e-05
At4g17170 46.6 1e-05
At1g73640 46.6 1e-05
Hs21361509 46.6 1e-05
Hs14777331 46.6 1e-05
At2g21880 46.2 1e-05
Hs7705963 46.2 1e-05
CE19650 46.2 1e-05
Hs4506371 46.2 1e-05
Hs7657492 46.2 1e-05
At4g19640 46.2 1e-05
At4g35860 46.2 2e-05
> SPBC1289.03c
Length=216
Score = 122 bits (306), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 55/66 (83%), Positives = 58/66 (87%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKYI TLGVEV PL F TNFG I FNVWDTAGQEK GGLRDGYYI+GQC I+MFDV
Sbjct 32 GEFEKKYIATLGVEVHPLHFHTNFGEICFNVWDTAGQEKLGGLRDGYYIQGQCGIIMFDV 91
Query 71 TSRVTY 76
TSR+TY
Sbjct 92 TSRITY 97
> Hs5453555
Length=216
Score = 122 bits (305), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/66 (84%), Positives = 59/66 (89%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKY+ TLGVEV PL F TN GPI FNVWDTAGQEKFGGLRDGYYI+ QCAI+MFDV
Sbjct 33 GEFEKKYVATLGVEVHPLVFHTNRGPIKFNVWDTAGQEKFGGLRDGYYIQAQCAIIMFDV 92
Query 71 TSRVTY 76
TSRVTY
Sbjct 93 TSRVTY 98
> 7292609
Length=216
Score = 119 bits (298), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 55/66 (83%), Positives = 59/66 (89%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKY+ TLGVEV PL F TN G I FNVWDTAGQEKFGGLRDGYYI+GQCA++MFDV
Sbjct 33 GEFEKKYVATLGVEVHPLIFHTNRGAIRFNVWDTAGQEKFGGLRDGYYIQGQCAVIMFDV 92
Query 71 TSRVTY 76
TSRVTY
Sbjct 93 TSRVTY 98
> YOR185c
Length=220
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 54/66 (81%), Positives = 58/66 (87%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKYI T+GVEV PL F TNFG I F+VWDTAGQEKFGGLRDGYYI QCAI+MFDV
Sbjct 36 GEFEKKYIATIGVEVHPLSFYTNFGEIKFDVWDTAGQEKFGGLRDGYYINAQCAIIMFDV 95
Query 71 TSRVTY 76
TSR+TY
Sbjct 96 TSRITY 101
> YLR293c
Length=219
Score = 118 bits (296), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 54/66 (81%), Positives = 58/66 (87%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKYI T+GVEV PL F TNFG I F+VWDTAGQEKFGGLRDGYYI QCAI+MFDV
Sbjct 35 GEFEKKYIATIGVEVHPLSFYTNFGEIKFDVWDTAGQEKFGGLRDGYYINAQCAIIMFDV 94
Query 71 TSRVTY 76
TSR+TY
Sbjct 95 TSRITY 100
> CE16194
Length=215
Score = 118 bits (296), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 55/66 (83%), Positives = 59/66 (89%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKY+ TLGVEV PL F TN G I FNVWDTAGQEKFGGLRDGYYI+GQCAI+MFDV
Sbjct 32 GEFEKKYVATLGVEVHPLVFHTNRGQIRFNVWDTAGQEKFGGLRDGYYIQGQCAIIMFDV 91
Query 71 TSRVTY 76
T+RVTY
Sbjct 92 TARVTY 97
> At5g55190
Length=221
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 52/66 (78%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKY PT+GVEV PL F TN G I F WDTAGQEKFGGLRDGYYI GQCAI+MFDV
Sbjct 36 GEFEKKYEPTIGVEVHPLDFFTNCGKIRFYCWDTAGQEKFGGLRDGYYIHGQCAIIMFDV 95
Query 71 TSRVTY 76
T+R+TY
Sbjct 96 TARLTY 101
> At5g20010
Length=221
Score = 112 bits (280), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 52/66 (78%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKY PT+GVEV PL F TN G I F WDTAGQEKFGGLRDGYYI GQCAI+MFDV
Sbjct 36 GEFEKKYEPTIGVEVHPLDFFTNCGKIRFYCWDTAGQEKFGGLRDGYYIHGQCAIIMFDV 95
Query 71 TSRVTY 76
T+R+TY
Sbjct 96 TARLTY 101
> At5g20020
Length=221
Score = 112 bits (280), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 52/66 (78%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKY PT+GVEV PL F TN G I F WDTAGQEKFGGLRDGYYI GQCAI+MFDV
Sbjct 36 GEFEKKYEPTIGVEVHPLDFFTNCGKIRFYCWDTAGQEKFGGLRDGYYIHGQCAIIMFDV 95
Query 71 TSRVTY 76
T+R+TY
Sbjct 96 TARLTY 101
> Hs20555668
Length=216
Score = 109 bits (273), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 50/66 (75%), Positives = 55/66 (83%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
GEFEKKY+ TLGVEV PL F TN GP+ FNVWDTAG EKF GLRDGYYI+ Q I++FDV
Sbjct 33 GEFEKKYVATLGVEVHPLVFHTNRGPVKFNVWDTAGLEKFSGLRDGYYIQAQSTIIVFDV 92
Query 71 TSRVTY 76
TSRVTY
Sbjct 93 TSRVTY 98
> Hs22056687
Length=109
Score = 108 bits (271), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 50/67 (74%), Positives = 55/67 (82%), Gaps = 0/67 (0%)
Query 10 AGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
GEFEKKY+ TLGVEV PL F TN GP+ FNVWDTAG EKF GLRDGYYI+ Q I++FD
Sbjct 32 TGEFEKKYVATLGVEVHPLVFHTNRGPVKFNVWDTAGLEKFSGLRDGYYIQAQSTIIVFD 91
Query 70 VTSRVTY 76
VTSRVTY
Sbjct 92 VTSRVTY 98
> At5g55080
Length=222
Score = 97.1 bits (240), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 44/67 (65%), Positives = 50/67 (74%), Gaps = 0/67 (0%)
Query 10 AGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
GEFE PTLGV++ PL F TN G I F WDTAGQEK+ GL+D YYI GQCAI+MFD
Sbjct 35 TGEFEHNTEPTLGVDIYPLDFFTNRGKIRFECWDTAGQEKYSGLKDAYYIHGQCAIIMFD 94
Query 70 VTSRVTY 76
VT+R TY
Sbjct 95 VTARHTY 101
> 7294292
Length=217
Score = 90.1 bits (222), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 41/67 (61%), Positives = 51/67 (76%), Gaps = 0/67 (0%)
Query 10 AGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
GEF+ +Y TLGVEV L F+TN G +VWDTAGQE++GGLRDGY+++ QCAI+MFD
Sbjct 32 TGEFKMQYNATLGVEVEQLLFNTNRGVFRIDVWDTAGQERYGGLRDGYFVQSQCAIIMFD 91
Query 70 VTSRVTY 76
V S TY
Sbjct 92 VASSNTY 98
> Hs20536248
Length=143
Score = 78.6 bits (192), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 39/53 (73%), Positives = 41/53 (77%), Gaps = 1/53 (1%)
Query 10 AGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQ 62
GEFEK I TLGVEV PL F TN GPI FNVWDTA +EKFG LRDGYYI+ Q
Sbjct 32 TGEFEK-CIATLGVEVHPLVFHTNRGPIKFNVWDTASREKFGRLRDGYYIQAQ 83
> ECU04g1560
Length=214
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/72 (50%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 5 VEAVRAGEFEKKYIPTLGVEVRPLRFSTNFGPIV-FNVWDTAGQEKFGGLRDGYYIKGQC 63
+ V G FEK Y T+G P+ F + G ++ FNVWDTAGQEK L+D YYI
Sbjct 24 INRVLDGRFEKNYNATVGAVNHPVTFLDDQGNVIKFNVWDTAGQEKKAVLKDVYYIGASG 83
Query 64 AIMMFDVTSRVT 75
AI FDVTSR+T
Sbjct 84 AIFFFDVTSRIT 95
> YOR089c
Length=210
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 0/62 (0%)
Query 10 AGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
+ +F + PT+G R + N + F +WDTAGQE+F L YY Q A++++D
Sbjct 29 SNDFAENKEPTIGAAFLTQRVTINEHTVKFEIWDTAGQERFASLAPMYYRNAQAALVVYD 88
Query 70 VT 71
VT
Sbjct 89 VT 90
> YNL093w
Length=220
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 39/67 (58%), Gaps = 0/67 (0%)
Query 10 AGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
+ +F++ PT+G R + + I F +WDTAGQE+F L YY Q A+++FD
Sbjct 34 SDDFKESKEPTIGAAFLTKRITRDGKVIKFEIWDTAGQERFAPLAPMYYRNAQAALVVFD 93
Query 70 VTSRVTY 76
VT+ ++
Sbjct 94 VTNEGSF 100
> Hs4758998
Length=333
Score = 50.8 bits (120), Expect = 7e-07, Method: Composition-based stats.
Identities = 24/59 (40%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 13 FEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 71
F++ Y T+GV+ RF P +WDTAGQEKF + YY Q I FD+T
Sbjct 148 FDRDYKATIGVDFEIERFEIAGIPYSLQIWDTAGQEKFKCIASAYYRGAQVIITAFDLT 206
> Hs5902040
Length=228
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/40 (50%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 37 IVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRVTY 76
I+ + WDTAGQE+F + YY K IM+FDV +VTY
Sbjct 70 ILVDFWDTAGQERFQSMHASYYHKAHACIMVFDVQRKVTY 109
> Hs5902038
Length=228
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 13 FEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 72
F+ + + T + + + + I+ + WDTAGQE+F + YY K IM+FD+
Sbjct 46 FQPQQLSTYALTLYKHTATVDGKTILVDFWDTAGQERFQSMHASYYHKAHACIMVFDIQR 105
Query 73 RVTY 76
+VTY
Sbjct 106 KVTY 109
> 7301214_2
Length=205
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 35/60 (58%), Gaps = 0/60 (0%)
Query 17 YIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRVTY 76
YI T+G++ + + + PI +WDTAGQE+F L YY ++M+DVT+ +Y
Sbjct 29 YISTIGIDFKQKLINLDGVPIKLQIWDTAGQERFRTLTTAYYRGAMGILLMYDVTNLESY 88
> CE29610
Length=220
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 13 FEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 72
F+ + T+GV+ R + + + +WDTAGQE+F L YY Q I ++DVTS
Sbjct 36 FDPEQAATIGVDFRVTSMAIDGNRVKLAIWDTAGQERFRTLTPSYYRGAQGVICVYDVTS 95
Query 73 RVTY 76
R ++
Sbjct 96 RSSF 99
> Hs19923481
Length=237
Score = 48.1 bits (113), Expect = 3e-06, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 37/74 (50%), Gaps = 0/74 (0%)
Query 3 SRVEAVRAGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQ 62
S ++ G F K Y T+GV+ + N + +WDTAGQE+F + YY Q
Sbjct 24 SMIQRYCKGIFTKDYKKTIGVDFLERQIQVNDEDVRLMLWDTAGQEEFDAITKAYYRGAQ 83
Query 63 CAIMMFDVTSRVTY 76
+++F T R ++
Sbjct 84 ACVLVFSTTDRGSF 97
> ECU09g1450
Length=203
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 37/68 (54%), Gaps = 0/68 (0%)
Query 9 RAGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMF 68
+ +F++ + T+GV+ N I+ N+WDTAGQE+F + YY +++F
Sbjct 30 KEDDFKQSLMTTIGVDTVSKSIVLNGKNIMLNIWDTAGQERFFSITKSYYRNADGILLIF 89
Query 69 DVTSRVTY 76
D++ T+
Sbjct 90 DISDERTF 97
> At1g43890
Length=212
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 19 PTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRVTY 76
PT+GV+ + + + +WDTAGQE+F L YY Q IM++DVT R T+
Sbjct 43 PTIGVDFKVKYLTIGEKKLKLAIWDTAGQERFRTLTSSYYRGAQGIIMVYDVTRRDTF 100
> At1g18200
Length=354
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 12 EFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 71
EF PT+GV+ I +WDTAGQE+F + YY A++++D+T
Sbjct 37 EFRLDSKPTIGVDFAYRNVRVGDKTIKAQIWDTAGQERFRAITSSYYRGALGALLIYDIT 96
Query 72 SRVTY 76
R+T+
Sbjct 97 RRITF 101
> Hs13994318
Length=259
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 22/58 (37%), Positives = 32/58 (55%), Gaps = 0/58 (0%)
Query 13 FEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
F+K Y T+GV+ RF P +WDTAGQE+F + YY Q I++F++
Sbjct 77 FDKNYKATIGVDFEMERFEVLGIPFSLQLWDTAGQERFKCIASTYYRGAQAIIIVFNL 134
> At3g09910
Length=205
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 19 PTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRVTY 76
PT+GV+ + + + +WDTAGQEKF L Y+ Q I+++DVT R T+
Sbjct 43 PTIGVDFKIKQMKVRGKRLKLTIWDTAGQEKFRTLTSSYFRGSQGIILVYDVTKRETF 100
> Hs4506375
Length=203
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 42/77 (54%), Gaps = 3/77 (3%)
Query 2 VSRVEAVRAGEFEKKYIPTLGVE--VRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYI 59
S V+ F K Y T+GV+ ++ L++S ++ + +WD AGQE+F + YY
Sbjct 21 TSLVQRYSQDSFSKHYKSTVGVDFALKVLQWS-DYEIVRLQLWDIAGQERFTSMTRLYYR 79
Query 60 KGQCAIMMFDVTSRVTY 76
++MFDVT+ T+
Sbjct 80 DASACVIMFDVTNATTF 96
> 7290172
Length=236
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNF----GPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIM 66
G F ++I T+G++ R R N I +WDTAGQE+F L +Y ++
Sbjct 40 GRFHTQFISTVGIDFREKRLLYNSRGRRHRIHLQIWDTAGQERFRSLTTAFYRDAMGFLL 99
Query 67 MFDVTSRVTY 76
+FD+TS ++
Sbjct 100 IFDLTSEKSF 109
> 7290171
Length=249
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNF----GPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIM 66
G F ++I T+G++ R R N I +WDTAGQE+F L +Y ++
Sbjct 53 GRFHTQFISTVGIDFREKRLLYNSRGRRHRIHLQIWDTAGQERFRSLTTAFYRDAMGFLL 112
Query 67 MFDVTSRVTY 76
+FD+TS ++
Sbjct 113 IFDLTSEKSF 122
> 7296421
Length=666
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 9 RAGEFEKKY-IPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMM 67
R G + Y + T+G++ R + + +WDTAGQE+F + YY +++
Sbjct 489 RDGRYVPSYFLSTVGIDFRNKVVVVDGTRVKLQIWDTAGQERFRSVTHAYYRDAHALLLL 548
Query 68 FDVTSRVTY 76
+DVT++ TY
Sbjct 549 YDVTNKTTY 557
> At5g47520
Length=221
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 12 EFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 71
EF T+GVE + + N I +WDTAGQE+F + YY A++++D++
Sbjct 38 EFYPNSKSTIGVEFQTQKMDINGKEIKAQIWDTAGQERFRAVTSAYYRGAVGALLVYDIS 97
Query 72 SRVTY 76
R T+
Sbjct 98 RRQTF 102
> Hs10880989
Length=206
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 13 FEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 72
F+ + T+GV+ + S + +WDTAGQE+F L YY Q I+++DVT
Sbjct 33 FDPELAATIGVDFKVKTISVDGNKAKLAIWDTAGQERFRTLTPSYYRGAQGVILVYDVTR 92
Query 73 RVTY 76
R T+
Sbjct 93 RDTF 96
> 7296691
Length=268
Score = 47.0 bits (110), Expect = 8e-06, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 38/74 (51%), Gaps = 0/74 (0%)
Query 3 SRVEAVRAGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQ 62
S ++ G F K Y T+GV+ + + + +WDTAGQE+F + YY Q
Sbjct 52 SMIQRYCKGIFTKDYKKTIGVDFLERQIEIDGEDVRIMLWDTAGQEEFDCITKAYYRGAQ 111
Query 63 CAIMMFDVTSRVTY 76
++++F T R ++
Sbjct 112 ASVLVFSTTDRASF 125
> Hs22047699
Length=217
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 37/72 (51%), Gaps = 0/72 (0%)
Query 5 VEAVRAGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCA 64
V+ ++G + + T+GV+ + + VWDTAGQE+F + YY A
Sbjct 34 VQHFKSGVYTETQQNTIGVDFTVRSLDIDGKKVKMQVWDTAGQERFRTITQSYYRSAHAA 93
Query 65 IMMFDVTSRVTY 76
I+ +D+T R T+
Sbjct 94 IIAYDLTRRSTF 105
> 7298153
Length=215
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 4/67 (5%)
Query 14 EKKYIP----TLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
EKK++ T+GVE + I +WDTAGQE+F + YY A+M++D
Sbjct 33 EKKFMANCPHTIGVEFGTRIIEVDDKKIKLQIWDTAGQERFRAVTRSYYRGAAGALMVYD 92
Query 70 VTSRVTY 76
+T R TY
Sbjct 93 ITRRSTY 99
> Hs22048268
Length=212
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 36/66 (54%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
EF +I T+GV+ + + + +WDTAGQE++ + YY + Q +++D+
Sbjct 31 NEFHSSHISTIGVDFKMKTIEVDGIKVRIQIWDTAGQERYQTITKQYYRRAQGIFLVYDI 90
Query 71 TSRVTY 76
+S +Y
Sbjct 91 SSERSY 96
> CE03777
Length=209
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 13 FEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTS 72
F +Y T+G + + + + +WDTAGQE+F L +Y C ++ FDVT+
Sbjct 34 FSNQYKATIGADFLTRDVNIDDRTVTLQIWDTAGQERFQSLGVAFYRGADCCVLAFDVTN 93
Query 73 RVTY 76
++
Sbjct 94 AASF 97
> At4g17170
Length=211
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 14 EKKYIP----TLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
+K++ P T+GVE + + PI +WDTAGQE F + YY A++++D
Sbjct 28 DKRFQPVHDLTIGVEFGARMITIDNKPIKLQIWDTAGQESFRSITRSYYRGAAGALLVYD 87
Query 70 VTSRVTY 76
+T R T+
Sbjct 88 ITRRETF 94
> At1g73640
Length=233
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 33/65 (50%), Gaps = 0/65 (0%)
Query 12 EFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 71
EF PT+GVE I +WDTAGQE+F + YY A++++D+T
Sbjct 37 EFRFDSKPTIGVEFAYRNVHVGDKIIKAQIWDTAGQERFRAITSSYYRGALGALLIYDIT 96
Query 72 SRVTY 76
R T+
Sbjct 97 RRTTF 101
> Hs21361509
Length=248
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 12 EFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 71
+F++ T+GVE + + +WDTAGQE+F + YY A++++D+T
Sbjct 67 KFKQDSNHTIGVEFGSRVVNVGGKTVKLQIWDTAGQERFRSVTRSYYRGAAGALLVYDIT 126
Query 72 SRVTY 76
SR TY
Sbjct 127 SRETY 131
> Hs14777331
Length=181
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 5 VEAVRAGEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCA 64
V+ + G F ++ T+GV+ + +WDTAGQE+F + YY A
Sbjct 4 VQRFKTGAFSERQGSTIGVDFTMKTLEIQGKRVKLQIWDTAGQERFRTITQSYYRSANGA 63
Query 65 IMMFDVTSRVTY 76
I+ +D+T R ++
Sbjct 64 ILAYDITKRSSF 75
> At2g21880
Length=230
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 12 EFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVT 71
+F K+Y T+G + + + +WDTAGQE+F L +Y C ++++DV
Sbjct 33 KFNKQYKATIGADFVTKELHIDEKSVTLQIWDTAGQERFQSLGAAFYRGADCCVLVYDVN 92
Query 72 S 72
+
Sbjct 93 N 93
> Hs7705963
Length=201
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 35/69 (50%), Gaps = 4/69 (5%)
Query 10 AGEFEKKYIPTLGVEV--RPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMM 67
+F+ + T+GVE R L F + +WDTAGQE+F LR +Y C ++
Sbjct 29 TNKFDSQAFHTIGVEFLNRDLEVDGRF--VTLQIWDTAGQERFKSLRTPFYRGADCCLLT 86
Query 68 FDVTSRVTY 76
F V R ++
Sbjct 87 FSVDDRQSF 95
> CE19650
Length=395
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 38/66 (57%), Gaps = 3/66 (4%)
Query 13 FEKKYIPTLGVE--VRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
F+ + T+GV+ V+ ++ N I +WDTAGQE+F + Y+ K ++MFDV
Sbjct 227 FKPLFNATIGVDFTVKTMKIPPNRA-IAMQLWDTAGQERFRSITKQYFRKADGVVLMFDV 285
Query 71 TSRVTY 76
TS ++
Sbjct 286 TSEQSF 291
> Hs4506371
Length=215
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 36/66 (54%), Gaps = 0/66 (0%)
Query 11 GEFEKKYIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDV 70
G+F + T+G + + F +WDTAGQE++ L YY Q AI+++D+
Sbjct 43 GQFHEYQESTIGAAFLTQSVCLDDTTVKFEIWDTAGQERYHSLAPMYYRGAQAAIVVYDI 102
Query 71 TSRVTY 76
T++ T+
Sbjct 103 TNQETF 108
> Hs7657492
Length=190
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 17 YIPTLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRVTY 76
+I T+G++ R + + +WDTAGQE+F + YY ++++DVT++ ++
Sbjct 27 FISTVGIDFRNKVLDVDGVKVKLQMWDTAGQERFRSVTHAYYRDAHALLLLYDVTNKASF 86
> At4g19640
Length=200
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 20 TLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFDVTSRVTY 76
T+G + N + F +WDTAGQE++ L YY AI++FDVT++ ++
Sbjct 42 TIGAAFFSQTLAVNDATVKFEIWDTAGQERYHSLAPMYYRGAAAAIIVFDVTNQASF 98
> At4g35860
Length=211
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 14 EKKYIP----TLGVEVRPLRFSTNFGPIVFNVWDTAGQEKFGGLRDGYYIKGQCAIMMFD 69
+K++ P T+GVE + + PI +WDTAGQE F + YY A++++D
Sbjct 28 DKRFQPVHDLTIGVEFGARMVTVDGRPIKLQIWDTAGQESFRSITRSYYRGAAGALLVYD 87
Query 70 VTSRVTY 76
+T R T+
Sbjct 88 ITRRETF 94
Lambda K H
0.325 0.142 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1178249128
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40