bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3485_orf2
Length=242
Score E
Sequences producing significant alignments: (Bits) Value
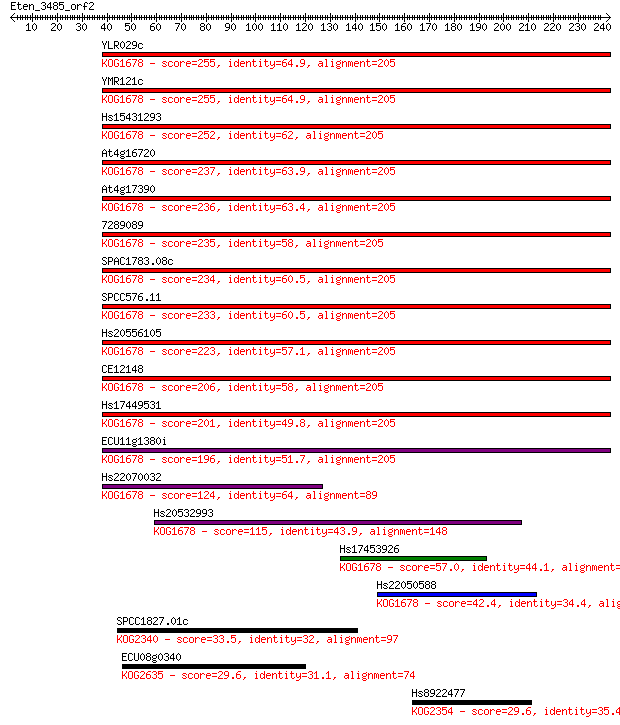
YLR029c 255 4e-68
YMR121c 255 5e-68
Hs15431293 252 4e-67
At4g16720 237 1e-62
At4g17390 236 2e-62
7289089 235 5e-62
SPAC1783.08c 234 2e-61
SPCC576.11 233 2e-61
Hs20556105 223 4e-58
CE12148 206 4e-53
Hs17449531 201 1e-51
ECU11g1380i 196 3e-50
Hs22070032 124 2e-28
Hs20532993 115 8e-26
Hs17453926 57.0 3e-08
Hs22050588 42.4 8e-04
SPCC1827.01c 33.5 0.45
ECU08g0340 29.6 5.2
Hs8922477 29.6 6.1
> YLR029c
Length=204
Score = 255 bits (652), Expect = 4e-68, Method: Compositional matrix adjust.
Identities = 133/205 (64%), Positives = 151/205 (73%), Gaps = 2/205 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKYLEEL ++KQSDVLRFL RVR WEYRQ +HR +RPTRPDKARRLGYK KQGFV
Sbjct 1 MGAYKYLEELQRKKQSDVLRFLQRVRVWEYRQKNVIHRAARPTRPDKARRLGYKAKQGFV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYRVRVRRG+RKR V KG YGKPTNQGV + K + +LR AE RV R NLR LNSY
Sbjct 61 IYRVRVRRGNRKRPVPKGATYGKPTNQGVNELKYQRSLRATAEERVGRR-AANLRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WV QD+ YKY+EVILVDP H AIR RY ++C KHRE RGLTA G+KSRG+N +G
Sbjct 120 WVNQDSTYKYFEVILVDPQHKAIRRDARYNWICDPVHKHREARGLTATGKKSRGIN-KGH 178
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
K + R WKR+ L L R+R
Sbjct 179 KFNNTKAGRRKTWKRQNTLSLWRYR 203
> YMR121c
Length=204
Score = 255 bits (652), Expect = 5e-68, Method: Compositional matrix adjust.
Identities = 133/205 (64%), Positives = 151/205 (73%), Gaps = 2/205 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKYLEEL ++KQSDVLRFL RVR WEYRQ +HR +RPTRPDKARRLGYK KQGFV
Sbjct 1 MGAYKYLEELERKKQSDVLRFLQRVRVWEYRQKNVIHRAARPTRPDKARRLGYKAKQGFV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYRVRVRRG+RKR V KG YGKPTNQGV + K + +LR AE RV R NLR LNSY
Sbjct 61 IYRVRVRRGNRKRPVPKGATYGKPTNQGVNELKYQRSLRATAEERVGRR-AANLRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WV QD+ YKY+EVILVDP H AIR RY ++C KHRE RGLTA G+KSRG+N +G
Sbjct 120 WVNQDSTYKYFEVILVDPQHKAIRRDARYNWICNPVHKHREARGLTATGKKSRGIN-KGH 178
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
K + R WKR+ L L R+R
Sbjct 179 KFNNTKAGRRKTWKRQNTLSLWRYR 203
> Hs15431293
Length=204
Score = 252 bits (644), Expect = 4e-67, Method: Compositional matrix adjust.
Identities = 127/205 (61%), Positives = 153/205 (74%), Gaps = 1/205 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKY++ELW++KQSDV+RFLLRVR W+YRQL A+HR RPTRPDKARRLGYK KQG+V
Sbjct 1 MGAYKYIQELWRKKQSDVMRFLLRVRCWQYRQLSALHRAPRPTRPDKARRLGYKAKQGYV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYR+RVRRG RKR V KG YGKP + GV + K +L+ VAE R R CG LR LNSY
Sbjct 61 IYRIRVRRGGRKRPVPKGATYGKPVHHGVNQLKFARSLQSVAEERAGRH-CGALRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WVG+D+ YK++EVIL+DP H AIR P +++ K KHRE+RGLT+AGRKSRGL
Sbjct 120 WVGEDSTYKFFEVILIDPFHKAIRRNPDTQWITKPVHKHREMRGLTSAGRKSRGLGKGHK 179
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
+ S RA W+RR LQL R+R
Sbjct 180 FHHTIGGSRRAAWRRRNTLQLHRYR 204
> At4g16720
Length=204
Score = 237 bits (605), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 131/205 (63%), Positives = 156/205 (76%), Gaps = 1/205 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKY+ ELW++KQSDV+RFL RVR WEYRQ P++ R RPTRPDKARRLGYK KQGFV
Sbjct 1 MGAYKYVSELWRKKQSDVMRFLQRVRCWEYRQQPSIVRLVRPTRPDKARRLGYKAKQGFV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
+YRVRVRRG RKR V KGIVYGKPTNQGV + K + + R VAE R R + G LR +NSY
Sbjct 61 VYRVRVRRGGRKRPVPKGIVYGKPTNQGVTQLKFQRSKRSVAEERAGRKL-GGLRVVNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
W+ +D+ YKYYE+ILVDP H A+RN PR ++C KHRELRGLT+ G+K+RGL +G
Sbjct 120 WLNEDSTYKYYEIILVDPAHNAVRNDPRINWICNPVHKHRELRGLTSEGKKNRGLRGKGH 179
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
N K PS RA WK+ L LRR+R
Sbjct 180 NNHKNRPSRRATWKKNNSLSLRRYR 204
> At4g17390
Length=204
Score = 236 bits (603), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 130/205 (63%), Positives = 156/205 (76%), Gaps = 1/205 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKY+ ELW++KQSDV+RFL RVR WEYRQ P++ R RPTRPDKARRLGYK KQGFV
Sbjct 1 MGAYKYVSELWRKKQSDVMRFLQRVRCWEYRQQPSIVRLVRPTRPDKARRLGYKAKQGFV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
+YRVRVRRG RKR V KGIVYGKPTNQGV + K + + R VAE R R + G LR +NSY
Sbjct 61 VYRVRVRRGGRKRPVPKGIVYGKPTNQGVTQLKFQRSKRSVAEERAGRKL-GGLRVVNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
W+ +D+ YKYYE+ILVDP H A+RN PR ++C KHRELRGLT+ G+K+RGL +G
Sbjct 120 WLNEDSTYKYYEIILVDPAHNAVRNDPRINWICNPVHKHRELRGLTSEGKKNRGLRGKGH 179
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
N K PS RA WK+ + LRR+R
Sbjct 180 NNHKNRPSRRATWKKNNSISLRRYR 204
> 7289089
Length=204
Score = 235 bits (600), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 119/206 (57%), Positives = 154/206 (74%), Gaps = 3/206 (1%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAY+Y++EL+++KQSDV+R+LLR+R W+YRQL +HR RPTRPDKARRLGY+ KQGFV
Sbjct 1 MGAYRYMQELYRKKQSDVMRYLLRIRVWQYRQLTKLHRSPRPTRPDKARRLGYRAKQGFV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYR+RVRRG RKR V KG YGKP + GV + K L+ +AE RV R + G LR LNSY
Sbjct 61 IYRIRVRRGGRKRPVPKGCTYGKPKSHGVNQLKPYRGLQSIAEERVGRRL-GGLRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
W+ QDA YKY+EVIL+D +H+AIR P+ ++CK KHRELRGLT+AG+ SRG+ +G
Sbjct 120 WIAQDASYKYFEVILIDTHHSAIRRDPKINWICKHVHKHRELRGLTSAGKSSRGIG-KGY 178
Query 218 KNSKRM-PSVRANWKRRQMLQLRRHR 242
+ S+ + S RA WKR+ + R R
Sbjct 179 RYSQTIGGSRRAAWKRKNREHMHRKR 204
> SPAC1783.08c
Length=201
Score = 234 bits (596), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 124/205 (60%), Positives = 143/205 (69%), Gaps = 4/205 (1%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKYLEEL K+KQSDV FL RVR WEYRQ+ +HR SRP+RPDKARRLGYK KQG+V
Sbjct 1 MGAYKYLEELAKKKQSDVNLFLSRVRAWEYRQMNVIHRASRPSRPDKARRLGYKAKQGYV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYR+RVRRG RKR V KG YGKP +QGV K + + R AE RV R C NLR LNSY
Sbjct 61 IYRIRVRRGGRKRPVPKGQTYGKPVHQGVNHLKYQRSARCTAEERVGR-YCSNLRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WV QDA YK++EVILVDP+H AIR PR ++ KHRE RGLT+ G+KSRG+ G
Sbjct 120 WVNQDATYKFFEVILVDPSHKAIRRDPRINWIVNPVHKHRESRGLTSIGKKSRGI---GK 176
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
+ A W R L LRR+R
Sbjct 177 GHRYNNSPQHATWLRHNTLSLRRYR 201
> SPCC576.11
Length=201
Score = 233 bits (595), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 124/205 (60%), Positives = 143/205 (69%), Gaps = 4/205 (1%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKYLEEL K+KQSDV FL RVR WEYRQ+ +HR SRP+RPDKARRLGYK KQG+V
Sbjct 1 MGAYKYLEELAKKKQSDVNLFLSRVRAWEYRQMNVIHRASRPSRPDKARRLGYKAKQGYV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYR+RVRRG RKR V KG YGKP +QGV K + + R AE RV R C NLR LNSY
Sbjct 61 IYRIRVRRGGRKRPVPKGQTYGKPVHQGVNHLKYQRSARCTAEERVGR-YCSNLRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WV QDA YK++EVILVDP+H AIR PR ++ KHRE RGLT+ G+KSRG+ G
Sbjct 120 WVNQDATYKFFEVILVDPSHKAIRRDPRINWIVNPVHKHRESRGLTSIGKKSRGI---GK 176
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
+ A W R L LRR+R
Sbjct 177 GHRFNNSPQHATWLRHNTLSLRRYR 201
> Hs20556105
Length=186
Score = 223 bits (567), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 117/205 (57%), Positives = 143/205 (69%), Gaps = 19/205 (9%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKY++ELW++KQSDV+RFLLRVR W+YRQL A+HR RPTRPDKARRLGYK KQG+V
Sbjct 1 MGAYKYIQELWRKKQSDVMRFLLRVRCWQYRQLSALHRAPRPTRPDKARRLGYKAKQGYV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYR+RVRRG R ++ K +L+ VAE RV R CG LR LNSY
Sbjct 61 IYRIRVRRGGRNQL------------------KFARSLQSVAEERVGRH-CGALRVLNSY 101
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WVG+D+ YK++EVIL+DP H AIR P +++ K KHRE+RGLT+AGRKSRGL
Sbjct 102 WVGEDSTYKFFEVILIDPFHKAIRRNPDTQWITKPVHKHREMRGLTSAGRKSRGLGKGHK 161
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
+ S RA W+RR LQL R+R
Sbjct 162 FHHTIGGSRRAAWRRRNTLQLHRYR 186
> CE12148
Length=204
Score = 206 bits (523), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 119/205 (58%), Positives = 147/205 (71%), Gaps = 1/205 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKY++E+W++KQSD LR+LLR+RTW YRQL AVHR RPTRP+KARRLGY+ KQGFV
Sbjct 1 MGAYKYMQEIWRKKQSDALRYLLRIRTWHYRQLSAVHRVPRPTRPEKARRLGYRAKQGFV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
+YRVRVRRG+RKR V KG YGKP GV + K + + VAE R R + G+LR LNSY
Sbjct 61 VYRVRVRRGNRKRPVCKGQTYGKPKTHGVNELKNAKSKQAVAEGRAGRRL-GSLRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WV +D+ YK+YEV+L+DP H AIR P +++ K KHRE RGLT+AGRKSRGL
Sbjct 120 WVAEDSTYKFYEVVLIDPFHKAIRRNPDTQWITKPVHKHREQRGLTSAGRKSRGLGKGWR 179
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
++ R S NWKR+ R R
Sbjct 180 FSATRGGSQAKNWKRKNTKVFHRKR 204
> Hs17449531
Length=204
Score = 201 bits (511), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 102/205 (49%), Positives = 136/205 (66%), Gaps = 1/205 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
M AYKY++E W++KQSDV+RFLLRV W+Y QL AVH PT PDKAR+LGYK KQG++
Sbjct 1 MDAYKYIQEQWRKKQSDVMRFLLRVCCWQYCQLSAVHGAPCPTWPDKARQLGYKAKQGYI 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
IYR+ V G +K + KG YGKP + GV + K +L+ +AE RV + CG LR LNSY
Sbjct 61 IYRIHVHHGGQKCPIPKGATYGKPVHHGVNQLKFARSLQSIAEERVGHN-CGALRVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WVG+D+ Y+++EVIL+DP + IR P + + K K RE+ GLT+AG++S GL
Sbjct 120 WVGEDSTYRFFEVILIDPFYEVIRRNPDTQSITKPVHKQREMCGLTSAGQRSCGLGKCHK 179
Query 218 KNSKRMPSVRANWKRRQMLQLRRHR 242
+ S A W+RR LQL R+
Sbjct 180 FHHTIDGSCWAAWRRRDTLQLHRYH 204
> ECU11g1380i
Length=204
Score = 196 bits (499), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 106/206 (51%), Positives = 139/206 (67%), Gaps = 3/206 (1%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGA YL E+ +RKQSDV+R+LLR+R EYRQ R +PT ++AR LGYK KQG+V
Sbjct 1 MGAVNYLREIHRRKQSDVMRYLLRIRVCEYRQRGECFRVEKPTFLERARTLGYKAKQGYV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSY 157
+Y RV++G+ KR G GK N G+ + K + + AE ++ C NL LNSY
Sbjct 61 LYIARVKKGNMKRNYHNGNTRGKCVNAGINQIKPSLRKQASAEM-IAGKKCSNLGVLNSY 119
Query 158 WVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSRGLNCRGA 217
WVGQDA YKYYEVI+VD +H AIRN P+ ++CK TMKHRE RGLT+A RKSRGL +G
Sbjct 120 WVGQDAAYKYYEVIMVDRDHQAIRNDPKINWICKGTMKHRECRGLTSASRKSRGLG-KGV 178
Query 218 K-NSKRMPSVRANWKRRQMLQLRRHR 242
K N + S+R+ W+RR L L+++R
Sbjct 179 KFNKSKGGSIRSCWRRRNTLVLQKYR 204
> Hs22070032
Length=90
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 57/89 (64%), Positives = 69/89 (77%), Gaps = 0/89 (0%)
Query 38 MGAYKYLEELWKRKQSDVLRFLLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFV 97
MGAYKY+ ELW++KQSDV+ FLLRVR W+YRQL A+HR RPTRPDK +LGYK KQG+V
Sbjct 1 MGAYKYIHELWRKKQSDVMSFLLRVRCWQYRQLSALHRAPRPTRPDKVHQLGYKIKQGYV 60
Query 98 IYRVRVRRGDRKRMVAKGIVYGKPTNQGV 126
IY +R+ R RK V KG YGKP + G+
Sbjct 61 IYGIRILRDGRKCSVPKGATYGKPVHHGI 89
> Hs20532993
Length=257
Score = 115 bits (288), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 65/148 (43%), Positives = 84/148 (56%), Gaps = 35/148 (23%)
Query 59 LLRVRTWEYRQLPAVHRCSRPTRPDKARRLGYKKKQGFVIYRVRVRRGDRKRMVAKGIVY 118
L VR QL A+HR R T+PDKA RLGYK KQG++IYR+ VRR
Sbjct 145 LPPVRAGTEWQLSALHRAPRSTQPDKACRLGYKAKQGYIIYRICVRR------------- 191
Query 119 GKPTNQGVRKQKAKMNLREVAERRVSRSVCGNLRCLNSYWVGQDAVYKYYEVILVDPNHA 178
+++A + CG LR LNSYWVG+D+ YK++EVIL+DP H
Sbjct 192 ---------EEQAGHH-------------CGALRVLNSYWVGEDSTYKFFEVILIDPFHK 229
Query 179 AIRNCPRYKYLCKSTMKHRELRGLTAAG 206
AIR P +++ K KHRE+RGLT+AG
Sbjct 230 AIRRNPDTQWITKPVHKHREMRGLTSAG 257
> Hs17453926
Length=156
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query 134 NLREVAERRVSRSVCGNLRCLNSYWVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKS 192
+L+ E R R G LR LNSYW G+ + YK++EVIL+DP H A+R P + + K+
Sbjct 89 SLQSFVEERAGRHF-GALRILNSYWAGEGSTYKFFEVILIDPFHKAMRRNPDTRRITKN 146
> Hs22050588
Length=333
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Query 149 GNLRCLNSYWVGQDAVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRK 208
G+LR LNSY G+D E +L+ H I+ P +++ + +KHRE+ GLT+ +
Sbjct 273 GSLRVLNSYGTGEDDPDPLGESLLI---HKVIKRNPDIQWVTREALKHREMCGLTSVATR 329
Query 209 SRGL 212
L
Sbjct 330 IMAL 333
> SPCC1827.01c
Length=652
Score = 33.5 bits (75), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 51/109 (46%), Gaps = 13/109 (11%)
Query 44 LEELWKRKQSDVLRFL------LRVRTW---EYRQLPAVHRCSRPTRPDKARRLGYKKKQ 94
L +L K QS +L ++ +RVR + E A+ S + +AR L Y+ +
Sbjct 500 LVQLKKYSQSGILVYIPSYFDFVRVRNYMDEEGINYSAISEYSSVSDMTRARNLFYQGRT 559
Query 95 GFVIYRVR---VRRGDRKRMVAKGIVYGKPTNQGVRKQKAKMNLREVAE 140
++Y R RR D R V ++YG PTN + +M +R ++E
Sbjct 560 NIMLYTERAHHFRRFDF-RGVKAVVMYGPPTNPQFYVELVRMPMRTISE 607
> ECU08g0340
Length=420
Score = 29.6 bits (65), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 8/82 (9%)
Query 46 ELWKRKQSDVLRFLLRVRTWE-------YRQLPAVHRCSRPTRPDKARRLGYKKKQGFVI 98
EL +R SD+ R L RVR+ E +QL ++ + S +K R L F++
Sbjct 143 ELQRRSVSDIDRELERVRSLEIEMRNESIQQLKSIVKTSSSPAKEKRRGLETSMSPVFIV 202
Query 99 YRVRVRR-GDRKRMVAKGIVYG 119
++ R++ D++ + G V G
Sbjct 203 FKERLKMVMDKENNIKSGEVQG 224
> Hs8922477
Length=708
Score = 29.6 bits (65), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Query 163 AVYKYYEVILVDPNHAAIRNCPRYKYLCKSTMKHRELRGLTAAGRKSR 210
A+Y+ E+ L P H ++ P + YL K+ KHRE AG S+
Sbjct 117 ALYRQGELHLT-PLHGILQLRPSFSYLDKADAKHREREAANEAGDSSQ 163
Lambda K H
0.325 0.137 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4883212382
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40