bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3480_orf2
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
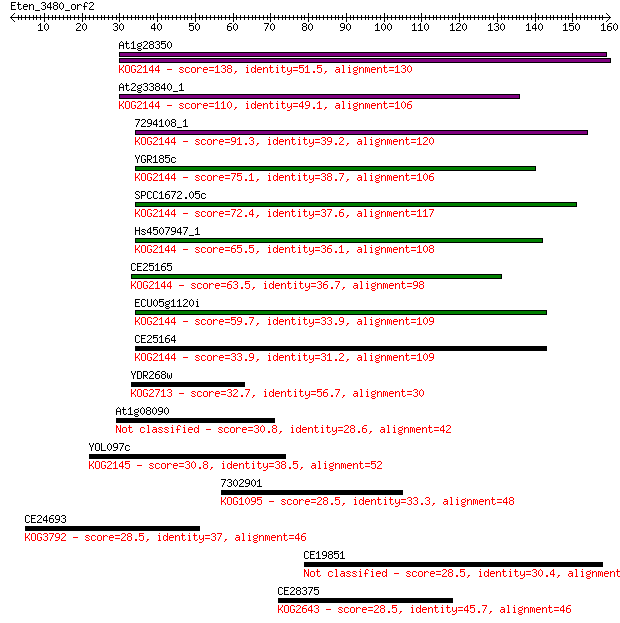
At1g28350 138 4e-33
At2g33840_1 110 1e-24
7294108_1 91.3 7e-19
YGR185c 75.1 5e-14
SPCC1672.05c 72.4 3e-13
Hs4507947_1 65.5 4e-11
CE25165 63.5 2e-10
ECU05g1120i 59.7 2e-09
CE25164 33.9 0.13
YDR268w 32.7 0.31
At1g08090 30.8 1.1
YOL097c 30.8 1.1
7302901 28.5 5.0
CE24693 28.5 5.1
CE19851 28.5 5.2
CE28375 28.5 5.9
> At1g28350
Length=866
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 66/130 (50%), Positives = 93/130 (71%), Gaps = 1/130 (0%)
Query 30 KDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP-DKEFHV 88
+ +KKMSKS+ SAIFMED EAEVN KI+KA+CPP V GNPC+ YV+ ++ P EF V
Sbjct 269 QGQKKMSKSDPSSAIFMEDEEAEVNVKIKKAYCPPDIVEGNPCLEYVKHIILPWFSEFTV 328
Query 89 QRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKL 148
+R + GGN FK+ E+ + +G+LHP DLK AL++ +N+ L+PVR HF+T+ AK L
Sbjct 329 ERDEKYGGNRTFKSFEDITTDYESGQLHPKDLKDALSKALNKILQPVRDHFKTNSRAKNL 388
Query 149 LQEISKYKIT 158
L+++ KI+
Sbjct 389 LKQVKVIKIS 398
Score = 116 bits (291), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 66/173 (38%), Positives = 91/173 (52%), Gaps = 44/173 (25%)
Query 30 KDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPD-KEFHV 88
+ ++KMSKS+ SAIFMED EA+VN KI KA+CPP V GNPC+ YV+ +V P EF V
Sbjct 695 QGQEKMSKSDPSSAIFMEDEEADVNEKISKAYCPPKTVEGNPCLEYVKYIVLPRFNEFKV 754
Query 89 QRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQL---------------- 132
+ +NGGN F + E+ + GELHP DLK AL + +N L
Sbjct 755 ESE-KNGGNKTFNSFEDIVADYETGELHPEDLKKALMKALNITLQFLASVQSSWNMKLVS 813
Query 133 --------------------------EPVRRHFQTDENAKKLLQEISKYKITR 159
+PVR HF+T+E AK LL+++ +++TR
Sbjct 814 FQSLLDVRVINFMYMNDMLLIVTVLSQPVRDHFKTNERAKNLLEQVKAFRVTR 866
> At2g33840_1
Length=379
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 52/107 (48%), Positives = 73/107 (68%), Gaps = 1/107 (0%)
Query 30 KDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP-DKEFHV 88
+ ++KMSKS+ SAIFMED EAEVN KI+KA+CPP V GNPC+ Y++ ++ P EF V
Sbjct 221 QGQEKMSKSDPLSAIFMEDEEAEVNVKIKKAYCPPKVVKGNPCLEYIKYIILPWFDEFTV 280
Query 89 QRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPV 135
+R E GGN +K+ E+ + +GELHP DLK L +N+ L+ +
Sbjct 281 ERNEEYGGNKTYKSFEDIAADYESGELHPGDLKKGLMNALNKILQVI 327
> 7294108_1
Length=365
Score = 91.3 bits (225), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 47/124 (37%), Positives = 75/124 (60%), Gaps = 9/124 (7%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP----DKEFHVQ 89
KMS S DS I + DS A V +K++KAFC P + N +++V+ ++F + F V
Sbjct 221 KMSSSEEDSKIDLLDSPANVKKKLKKAFCEPGNIADNGLLSFVKHVLFSLFKEGEGFEVN 280
Query 90 RTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKLL 149
R E+GG++ F E+ ++ +A +LHP DLKA + + INR L+P+R+ F+ E L
Sbjct 281 REAEHGGDVTFLKYEDLEKYYAEDKLHPGDLKATVEKYINRLLDPIRKAFENPE-----L 335
Query 150 QEIS 153
Q++S
Sbjct 336 QKLS 339
> YGR185c
Length=394
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 63/115 (54%), Gaps = 9/115 (7%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKE-------- 85
KMS S+ +S I + + +V +KI AFC P V N +++V+ ++ P +E
Sbjct 227 KMSASDPNSKIDLLEEPKQVKKKINSAFCSPGNVEENGLLSFVQYVIAPIQELKFGTNHF 286
Query 86 -FHVQRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHF 139
F + R + GG I +K+ EE K +F +L P DLK +A IN LEP+R+ F
Sbjct 287 EFFIDRPEKFGGPITYKSFEEMKLAFKEEKLSPPDLKIGVADAINELLEPIRQEF 341
> SPCC1672.05c
Length=401
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 66/124 (53%), Gaps = 7/124 (5%)
Query 34 KMSKSNADSA-IFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP------DKEF 86
KMS S A +A I + D VN+KIR AFC P V N C+A+++ + FP + EF
Sbjct 220 KMSSSAAANAKIDILDDPKTVNKKIRSAFCEPGVVEENGCLAFLKYVSFPAMELRGETEF 279
Query 87 HVQRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAK 146
+ R + GG +VFKT EE ++++ L P DLK L +N L V+ + +
Sbjct 280 VIDRPEQFGGRMVFKTYEEVEKAYKDLTLSPQDLKLGLESSVNTLLAGVQEQLKQYPDLD 339
Query 147 KLLQ 150
+L+
Sbjct 340 SILK 343
> Hs4507947_1
Length=356
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/109 (35%), Positives = 66/109 (60%), Gaps = 1/109 (0%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDK-EFHVQRTL 92
KMS S +S I + D + +V +K++KAFC P V N +++++ ++FP K EF + R
Sbjct 222 KMSSSEEESKIDLLDRKEDVKKKLKKAFCEPGNVENNGVLSFIKHVLFPLKSEFVILRDE 281
Query 93 ENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQT 141
+ GGN + + ++ FAA +HP DLK ++ +N+ L+P+R F T
Sbjct 282 KWGGNKTYTAYVDLEKDFAAEVVHPGDLKNSVEVALNKLLDPIREKFNT 330
> CE25165
Length=275
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 55/108 (50%), Gaps = 10/108 (9%)
Query 33 KKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP---DKEFHVQ 89
+KMS S D + D+ ++ KI ++FC P + GN + E +VFP H+Q
Sbjct 78 QKMSCSIPDFLLDPLDTPKQLKTKIARSFCEPQNLDGNVAMQLAEHIVFPILNGAALHIQ 137
Query 90 RTLENGGNIVFKTAEEFKRSFAAG-------ELHPADLKAALAREINR 130
R +NGG++ ++ ++ F G LHPADLK A+ INR
Sbjct 138 RAPDNGGDVEVTNYKQLEQEFVIGTKNEQQFPLHPADLKNAVVGVINR 185
> ECU05g1120i
Length=337
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/112 (33%), Positives = 56/112 (50%), Gaps = 4/112 (3%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKEFH---VQR 90
KMS S+ S I + DS+ + RKIRK FC N + +VFP + V+
Sbjct 218 KMSSSDDLSKIDLMDSKEAIWRKIRKCFCEEGN-KDNGLMMIFSHIVFPILQLKGECVRI 276
Query 91 TLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTD 142
T +G + F+ +EF+ F +HP DLK+ AR I+ + P+R + D
Sbjct 277 TDRDGREMAFEKYQEFEEEFVRKSIHPGDLKSNAARLIDEIIRPIREEMEKD 328
> CE25164
Length=540
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 34/109 (31%), Positives = 46/109 (42%), Gaps = 5/109 (4%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKEFHVQRTLE 93
KMS S DS I + D +V KI A C Q N +++ ++FP V
Sbjct 233 KMSSSEEDSKIDVLDEPEKVRSKIMGAACSRDQ-PDNGVLSFYNFVLFPI----VSPNAI 287
Query 94 NGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTD 142
N F + KR++ G+L LK LA + LE VR TD
Sbjct 288 EIENQQFFDIDSLKRAYLDGKLDENTLKTFLADFLVNLLEKVRVRCDTD 336
> YDR268w
Length=379
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 20/32 (62%), Gaps = 2/32 (6%)
Query 33 KKMSKS--NADSAIFMEDSEAEVNRKIRKAFC 62
KKMSKS N DS IF+ D + +KIRKA
Sbjct 243 KKMSKSDPNHDSVIFLNDEPKAIQKKIRKALT 274
> At1g08090
Length=530
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 12/42 (28%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 29 WKDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGN 70
W +++K + S F E++++E R++R A PP N
Sbjct 488 WNEQEKQKNMHQGSLRFAENAKSEGGRRVRSAATPPENTPNN 529
> YOL097c
Length=432
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 27/60 (45%), Gaps = 8/60 (13%)
Query 22 TRCFRDCWKDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVH--------GNPCV 73
+R F KMS S+ +AIFM D+ ++ +KI K QV GNP V
Sbjct 283 SRFFPALQGSTTKMSASDDTTAIFMTDTPKQIQKKINKYAFSGGQVSADLHRELGGNPDV 342
> 7302901
Length=1701
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 21/48 (43%), Gaps = 3/48 (6%)
Query 57 IRKAFCPPAQVHGNPCVAYVERLVFPDKEFHVQRTLENGGNIVFKTAE 104
+ K F PP VHGNP Y V F + + +N G+I E
Sbjct 582 VSKTFNPPPSVHGNPDSPYRNSCVV---RFFIHQFGKNPGSINLSVVE 626
> CE24693
Length=561
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 5 GSTATKSRENTNLSFCLTRCFRDCWKDRKKMSKSNADSAIFMEDSE 50
ST++ REN S L + FR W RKK++ S +IF +++E
Sbjct 2 ASTSSAWRENAQKSENLAKKFRKIWIFRKKLAFSRQKCSIFDKNTE 47
> CE19851
Length=229
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 39/85 (45%), Gaps = 12/85 (14%)
Query 79 LVFPDKEFHVQRTLENG------GNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQL 132
L+ D + HV + + G F++ E+ K F ++ D+KA LA +
Sbjct 72 LLVEDTKLHVNKAFLSYHSEFFRGLFEFESEEKQKNEFKISDVSADDMKAFLAI-----I 126
Query 133 EPVRRHFQTDENAKKLLQEISKYKI 157
P F TD NAKKLL+ ++ +
Sbjct 127 HP-GNDFPTDSNAKKLLELADRFMV 150
> CE28375
Length=561
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 26/56 (46%), Gaps = 10/56 (17%)
Query 72 CVAYVERLVF---PDKEFHVQRTL------ENGGNI-VFKTAEEFKRSFAAGELHP 117
+ Y +RL PDK F TL E+ G VF T E+F RSF G + P
Sbjct 192 IIEYEDRLRLYSTPDKIFRYFATLKIIDPNEDSGRFEVFMTPEDFLRSFTPGVMQP 247
Lambda K H
0.320 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40