bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
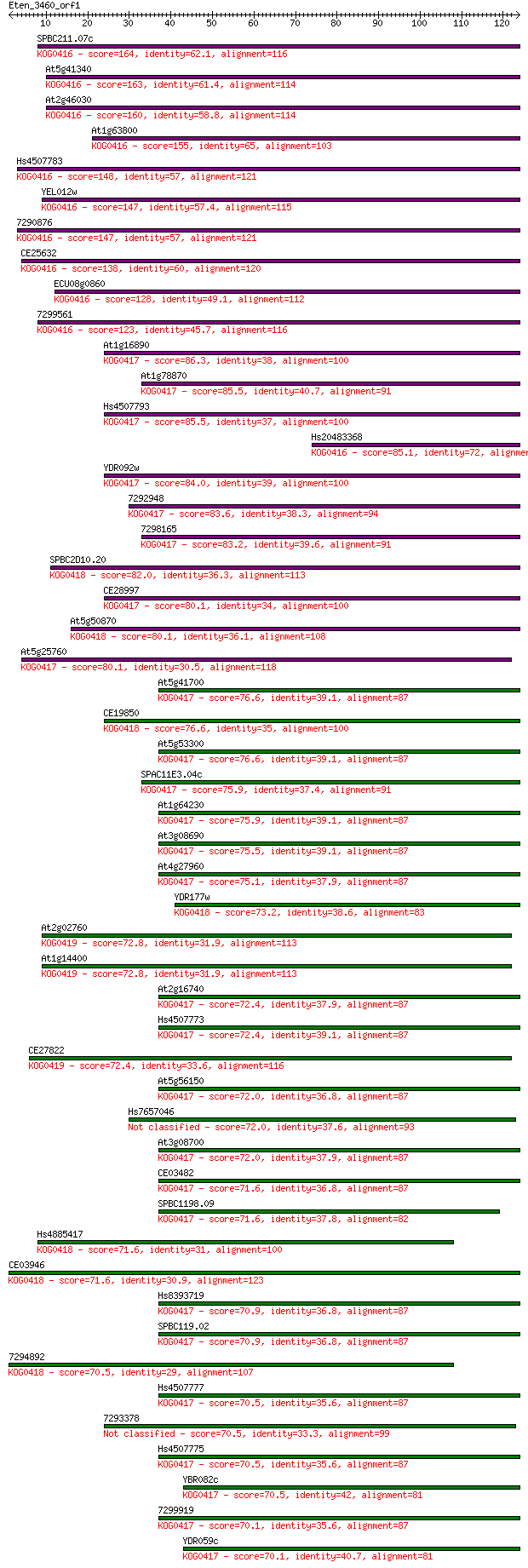
Query= Eten_3460_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
SPBC211.07c 164 5e-41
At5g41340 163 7e-41
At2g46030 160 4e-40
At1g63800 155 1e-38
Hs4507783 148 3e-36
YEL012w 147 3e-36
7290876 147 6e-36
CE25632 138 2e-33
ECU08g0860 128 2e-30
7299561 123 1e-28
At1g16890 86.3 1e-17
At1g78870 85.5 2e-17
Hs4507793 85.5 2e-17
Hs20483368 85.1 3e-17
YDR092w 84.0 7e-17
7292948 83.6 9e-17
7298165 83.2 1e-16
SPBC2D10.20 82.0 2e-16
CE28997 80.1 9e-16
At5g50870 80.1 1e-15
At5g25760 80.1 1e-15
At5g41700 76.6 1e-14
CE19850 76.6 1e-14
At5g53300 76.6 1e-14
SPAC11E3.04c 75.9 2e-14
At1g64230 75.9 2e-14
At3g08690 75.5 3e-14
At4g27960 75.1 3e-14
YDR177w 73.2 1e-13
At2g02760 72.8 1e-13
At1g14400 72.8 2e-13
At2g16740 72.4 2e-13
Hs4507773 72.4 2e-13
CE27822 72.4 2e-13
At5g56150 72.0 2e-13
Hs7657046 72.0 3e-13
At3g08700 72.0 3e-13
CE03482 71.6 3e-13
SPBC1198.09 71.6 3e-13
Hs4885417 71.6 3e-13
CE03946 71.6 4e-13
Hs8393719 70.9 5e-13
SPBC119.02 70.9 6e-13
7294892 70.5 7e-13
Hs4507777 70.5 7e-13
7293378 70.5 7e-13
Hs4507775 70.5 7e-13
YBR082c 70.5 8e-13
7299919 70.1 9e-13
YDR059c 70.1 1e-12
> SPBC211.07c
Length=184
Score = 164 bits (414), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 72/118 (61%), Positives = 93/118 (78%), Gaps = 2/118 (1%)
Query 8 ISSSRK--QCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYP 65
+SS R+ + D KLLM+ Y++ L N N Q+F V FHGP ET Y GG+W+VHV LP +YP
Sbjct 1 MSSPRRRIETDVMKLLMSDYEVTLVNDNMQEFYVRFHGPSETPYSGGIWKVHVELPSEYP 60
Query 66 FSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
+ SPSIGF+N+I HPN+DE SGSVCLDVINQTW+P++ ++N+F+VFLPQLL YPN D
Sbjct 61 WKSPSIGFVNRIFHPNIDELSGSVCLDVINQTWSPMFDMINIFEVFLPQLLRYPNASD 118
> At5g41340
Length=187
Score = 163 bits (413), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 70/114 (61%), Positives = 90/114 (78%), Gaps = 0/114 (0%)
Query 10 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSP 69
S R++ D KL+M+ Y +E N Q+F V F+GPK+++Y+GGVW++ V LPD YP+ SP
Sbjct 5 SKRREMDMMKLMMSDYKVETINDGMQEFYVEFNGPKDSLYQGGVWKIRVELPDAYPYKSP 64
Query 70 SIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
S+GF+ KI HPNVDE SGSVCLDVINQTW+P++ LVNVF+ FLPQLL YPNP D
Sbjct 65 SVGFITKIYHPNVDELSGSVCLDVINQTWSPMFDLVNVFETFLPQLLLYPNPSD 118
> At2g46030
Length=183
Score = 160 bits (406), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 67/114 (58%), Positives = 91/114 (79%), Gaps = 0/114 (0%)
Query 10 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSP 69
S R++ D KL+M+ Y ++ N + Q F V FHGP +++Y+GGVW++ V LP+ YP+ SP
Sbjct 5 SKRREMDMMKLMMSDYKVDTVNDDLQMFYVTFHGPTDSLYQGGVWKIKVELPEAYPYKSP 64
Query 70 SIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
S+GF+NKI HPNVDE+SG+VCLDVINQTW+P++ L+NVF+ FLPQLL YPNP D
Sbjct 65 SVGFVNKIYHPNVDESSGAVCLDVINQTWSPMFDLINVFESFLPQLLLYPNPSD 118
> At1g63800
Length=170
Score = 155 bits (393), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 67/103 (65%), Positives = 83/103 (80%), Gaps = 0/103 (0%)
Query 21 LMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHP 80
+M+ Y +E+ N Q+F V F GPK++IYEGGVW++ V LPD YP+ SPS+GF+ KI HP
Sbjct 1 MMSDYKVEMINDGMQEFFVEFSGPKDSIYEGGVWKIRVELPDAYPYKSPSVGFITKIYHP 60
Query 81 NVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
NVDE SGSVCLDVINQTW+P++ LVNVF+ FLPQLL YPNP D
Sbjct 61 NVDEMSGSVCLDVINQTWSPMFDLVNVFETFLPQLLLYPNPSD 103
> Hs4507783
Length=183
Score = 148 bits (373), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 69/121 (57%), Positives = 85/121 (70%), Gaps = 1/121 (0%)
Query 3 MAQTQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 62
M+ R D KL+ + +++ + +F V F+GP+ T YEGGVW+V V LPD
Sbjct 1 MSSPSPGKRRMDTDVVKLIESKHEVTILG-GLNEFVVKFYGPQGTPYEGGVWKVRVDLPD 59
Query 63 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 122
YPF SPSIGF+NKI HPN+DEASG+VCLDVINQTWT LY L N+F+ FLPQLL YPNP
Sbjct 60 KYPFKSPSIGFMNKIFHPNIDEASGTVCLDVINQTWTALYDLTNIFESFLPQLLAYPNPI 119
Query 123 D 123
D
Sbjct 120 D 120
> YEL012w
Length=218
Score = 147 bits (372), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 66/115 (57%), Positives = 86/115 (74%), Gaps = 0/115 (0%)
Query 9 SSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSS 68
S R + D KLLM+ + ++L N + Q+F+V F GPK+T YE GVWR+HV LPD+YP+ S
Sbjct 4 SKRRIETDVMKLLMSDHQVDLINDSMQEFHVKFLGPKDTPYENGVWRLHVELPDNYPYKS 63
Query 69 PSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
PSIGF+NKI HPN+D ASGS+CLDVIN TW+PLY L+N+ + +P LL PN D
Sbjct 64 PSIGFVNKIFHPNIDIASGSICLDVINSTWSPLYDLINIVEWMIPGLLKEPNGSD 118
> 7290876
Length=183
Score = 147 bits (370), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 69/121 (57%), Positives = 86/121 (71%), Gaps = 1/121 (0%)
Query 3 MAQTQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 62
M+ R D KL+ + +++ + +F+V F GP ET YEGGVW+V V LPD
Sbjct 1 MSSPSAGKRRMDNDVIKLIESKHEVTILG-GLNEFHVKFFGPTETPYEGGVWKVRVYLPD 59
Query 63 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 122
+YPF SPSIGF+NKI HPN+DE+SG+VCLDVINQ WT LY L N+F+ FLPQLLTYPNP
Sbjct 60 NYPFKSPSIGFVNKIYHPNIDESSGTVCLDVINQAWTALYDLSNIFESFLPQLLTYPNPV 119
Query 123 D 123
D
Sbjct 120 D 120
> CE25632
Length=241
Score = 138 bits (348), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 72/153 (47%), Positives = 86/153 (56%), Gaps = 34/153 (22%)
Query 4 AQTQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKE----------------- 46
+ T I R CD KL+ +++++ N +F V FHGPK+
Sbjct 3 SATAIGKRRIDCDVVKLISHNHEVQIVN-GCSEFIVRFHGPKDRNFSPKNFNFRQVLPIF 61
Query 47 ------TIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ---- 96
YE GVWR+ V +PD YPF SPSIGFLNKI HPN+DEASG+VCLDVINQ
Sbjct 62 RTKILAAAYENGVWRIRVDMPDKYPFKSPSIGFLNKIFHPNIDEASGTVCLDVINQVGIG 121
Query 97 ------TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
WT LY L N+FD FLPQLLTYPN D
Sbjct 122 GRSVWKAWTALYDLTNIFDTFLPQLLTYPNAAD 154
> ECU08g0860
Length=171
Score = 128 bits (322), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 55/112 (49%), Positives = 78/112 (69%), Gaps = 0/112 (0%)
Query 12 RKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSI 71
R + + KL A Y++ + V+ +GP+++ Y G + V++ LPD+YPF SPSI
Sbjct 7 RIKNEIQKLYNANYEVRMIEGKQTQMEVLINGPEDSPYSGAKYIVNILLPDNYPFKSPSI 66
Query 72 GFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
GF +I HPNVDEASGS+CLDV+NQ W+P+Y L+N+ D+ LPQLL+YPN D
Sbjct 67 GFTTRIFHPNVDEASGSICLDVLNQIWSPIYDLLNIVDILLPQLLSYPNAAD 118
> 7299561
Length=206
Score = 123 bits (308), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 53/116 (45%), Positives = 78/116 (67%), Gaps = 1/116 (0%)
Query 8 ISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFS 67
++ R D +LL +GY +++ T + NV GP + YEGG+W V+VT+P DYP +
Sbjct 13 MAGRRLDRDVNRLLASGYRTTVDDDMT-NLNVCLEGPLGSAYEGGIWTVNVTMPQDYPLT 71
Query 68 SPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
+P + F+ KILHPN++ +G VC++V+ Q W+ Y LVN+F+ FLPQLL YPNP D
Sbjct 72 APRVRFVTKILHPNIEFITGLVCMNVLKQAWSSSYDLVNIFETFLPQLLRYPNPHD 127
> At1g16890
Length=153
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 38/100 (38%), Positives = 63/100 (63%), Gaps = 2/100 (2%)
Query 24 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 83
G + +N + FNV+ GP ++ YEGGV+++ + LP++YP ++P + FL KI HPN+D
Sbjct 24 GISASPSEENMRYFNVMILGPTQSPYEGGVFKLELFLPEEYPMAAPKVRFLTKIYHPNID 83
Query 84 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
+ G +CLD++ W+P + V + + LL+ PNP D
Sbjct 84 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDD 121
> At1g78870
Length=153
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 37/91 (40%), Positives = 60/91 (65%), Gaps = 2/91 (2%)
Query 33 NTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLD 92
N + FNV+ GP ++ YEGGV+++ + LP++YP ++P + FL KI HPN+D+ G +CLD
Sbjct 33 NMRYFNVMILGPTQSPYEGGVFKLELFLPEEYPMAAPKVRFLTKIYHPNIDKL-GRICLD 91
Query 93 VINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
++ W+P + V + + LL+ PNP D
Sbjct 92 ILKDKWSPALQIRTVL-LSIQALLSAPNPDD 121
> Hs4507793
Length=152
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 63/100 (63%), Gaps = 2/100 (2%)
Query 24 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 83
G E + N + F+VV GP+++ +EGG +++ + LP++YP ++P + F+ KI HPNVD
Sbjct 22 GIKAEPDESNARYFHVVIAGPQDSPFEGGTFKLELFLPEEYPMAAPKVRFMTKIYHPNVD 81
Query 84 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
+ G +CLD++ W+P + V + + LL+ PNP D
Sbjct 82 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDD 119
> Hs20483368
Length=113
Score = 85.1 bits (209), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 36/50 (72%), Positives = 42/50 (84%), Gaps = 0/50 (0%)
Query 74 LNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
++KI HPN D+ASG+VCLDVINQTWT LY L N+F+ FLPQLL YPNP D
Sbjct 1 MHKIFHPNTDDASGTVCLDVINQTWTALYDLTNIFESFLPQLLAYPNPID 50
> YDR092w
Length=153
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 58/100 (58%), Gaps = 2/100 (2%)
Query 24 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 83
G E ++ N + F V GP+++ YE G++ + + LPDDYP +P + FL KI HPN+D
Sbjct 22 GITAEPHDDNLRYFQVTIEGPEQSPYEDGIFELELYLPDDYPMEAPKVRFLTKIYHPNID 81
Query 84 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
G +CLDV+ W+P + V + + LL PNP D
Sbjct 82 RL-GRICLDVLKTNWSPALQIRTVL-LSIQALLASPNPND 119
> 7292948
Length=151
Score = 83.6 bits (205), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 60/94 (63%), Gaps = 2/94 (2%)
Query 30 NNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSV 89
+ N + F+V+ GP ++ +EGGV+++ + LP+DYP S+P + F+ KI HPN+D G +
Sbjct 28 DENNARYFHVIVTGPNDSPFEGGVFKLELFLPEDYPMSAPKVRFITKIYHPNIDRL-GRI 86
Query 90 CLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
CLDV+ W+P + + + + LL+ PNP D
Sbjct 87 CLDVLKDKWSPALQIRTIL-LSIQALLSAPNPDD 119
> 7298165
Length=151
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/91 (39%), Positives = 58/91 (63%), Gaps = 2/91 (2%)
Query 33 NTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLD 92
N + F+V+ GPK++ +EGG +++ + LP+DYP +P + FL KI HPN+D G +CLD
Sbjct 31 NARYFHVLVTGPKDSPFEGGNFKLELFLPEDYPMKAPKVRFLTKIFHPNIDRV-GRICLD 89
Query 93 VINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
++ W+P + V + + LL+ PNP D
Sbjct 90 ILKDKWSPALQIRTVL-LSIQALLSAPNPDD 119
> SPBC2D10.20
Length=217
Score = 82.0 bits (201), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 41/116 (35%), Positives = 64/116 (55%), Gaps = 4/116 (3%)
Query 11 SRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPS 70
+++ D + AG + N + +F GP+ T YEGG + V + +P DYPF P
Sbjct 10 AKELADVQQDKQAGIQVWTINDDISHLKGMFRGPEGTPYEGGYFVVDIEIPIDYPFRPPK 69
Query 71 IGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYP---NPQD 123
+ F KI HPNV +G++CLD++ W+P+Y++ + + L LL P NPQD
Sbjct 70 MNFDTKIYHPNVSSQTGAICLDILKDQWSPVYTMKSAL-ISLQSLLCTPEPSNPQD 124
> CE28997
Length=185
Score = 80.1 bits (196), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 34/100 (34%), Positives = 62/100 (62%), Gaps = 2/100 (2%)
Query 24 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 83
G + N + F+V+ GP ++ + GGV+++ + LP++YP ++P + F+ KI HPN+D
Sbjct 23 GISANPDESNARYFHVMIAGPDDSPFAGGVFKLELFLPEEYPMAAPKVRFMTKIYHPNID 82
Query 84 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
+ G +CLD++ W+P + V + + LL+ PNP+D
Sbjct 83 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPED 120
> At5g50870
Length=192
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 60/119 (50%), Gaps = 12/119 (10%)
Query 16 DFAKLLMAGYDLELN-----------NKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDY 64
DF+++ D E N + N GP T YEGG +++ +T+PD Y
Sbjct 3 DFSRIQKELQDCERNQDSSGIRVCPKSDNLTRLTGTIPGPIGTPYEGGTFQIDITMPDGY 62
Query 65 PFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
PF P + F K+ HPN+ SG++CLD++ W+P +L V + LL+ P P+D
Sbjct 63 PFEPPKMQFSTKVWHPNISSQSGAICLDILKDQWSPALTLKTAL-VSIQALLSAPEPKD 120
> At5g25760
Length=157
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 64/118 (54%), Gaps = 1/118 (0%)
Query 4 AQTQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDD 63
++ ++ K+ K+ L ++ N + + GP ET YEGGV+++ ++P+
Sbjct 4 SRARLFKEYKEVQREKVADPDIQLICDDTNIFKWTALIKGPSETPYEGGVFQLAFSVPEP 63
Query 64 YPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNP 121
YP P + FL KI HPNV +G +CLD++ W+P ++L +V + L+ +P P
Sbjct 64 YPLQPPQVRFLTKIFHPNVHFKTGEICLDILKNAWSPAWTLQSVCRAII-ALMAHPEP 120
> At5g41700
Length=148
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP E+ Y GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ +
Sbjct 33 WQATIMGPAESPYSGGVFLVTIHFPPDYPFKPPKVAFRTKVFHPNIN-SNGSICLDILKE 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLTDPNPDD 117
> CE19850
Length=199
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 56/100 (56%), Gaps = 1/100 (1%)
Query 24 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 83
G +E+ N+N + GP +T Y GG++ + + +PD YPFS P++ F KI HPNV
Sbjct 26 GIMIEILNENLTEIKGHIRGPPDTPYAGGMFDLDIKIPDQYPFSPPNVKFSTKIWHPNVS 85
Query 84 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
+G +CLD++ W +L V + + L+ P P+D
Sbjct 86 SQTGVICLDILKDQWAASLTLRTVL-LSIQALMCTPEPKD 124
> At5g53300
Length=148
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP E+ Y GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ +
Sbjct 33 WQATIMGPSESPYAGGVFLVTIHFPPDYPFKPPKVAFRTKVFHPNIN-SNGSICLDILKE 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLTDPNPDD 117
> SPAC11E3.04c
Length=148
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 54/91 (59%), Gaps = 2/91 (2%)
Query 33 NTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLD 92
N + F + GP+++ YEGG + + + LPD+YP P++ FL KI HPNVD+ G +CL
Sbjct 30 NLRYFKITMEGPQQSAYEGGKFHLELFLPDEYPMMPPNVRFLTKIYHPNVDKL-GRICLS 88
Query 93 VINQTWTPLYSLVNVFDVFLPQLLTYPNPQD 123
+ + W+P + V + + L+ PNP D
Sbjct 89 TLKKDWSPALQIRTVL-LSIQALMGAPNPDD 118
> At1g64230
Length=173
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y GGV+ V + P DYPF P + F K+ HPNV+ ++GS+CLD++ +
Sbjct 58 WQATIMGPSDSPYSGGVFLVTIHFPPDYPFKPPKVAFRTKVFHPNVN-SNGSICLDILKE 116
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 117 QWSPALTISKVL-LSICSLLTDPNPDD 142
> At3g08690
Length=148
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP E+ Y GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ +
Sbjct 33 WQATIMGPPESPYAGGVFLVSIHFPPDYPFKPPKVSFKTKVYHPNIN-SNGSICLDILKE 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLTDPNPDD 117
> At4g27960
Length=148
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ +
Sbjct 33 WQATIMGPSDSPYSGGVFLVTIHFPPDYPFKPPKVAFRTKVFHPNIN-SNGSICLDILKE 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLTDPNPDD 117
> YDR177w
Length=215
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 49/83 (59%), Gaps = 1/83 (1%)
Query 41 FHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTP 100
F GP T YEGG + V + +P +YPF P + F K+ HPN+ +G++CLD++ W+P
Sbjct 39 FLGPPGTPYEGGKFVVDIEVPMEYPFKPPKMQFDTKVYHPNISSVTGAICLDILKNAWSP 98
Query 101 LYSLVNVFDVFLPQLLTYPNPQD 123
+ +L + + L LL P P D
Sbjct 99 VITLKSAL-ISLQALLQSPEPND 120
> At2g02760
Length=152
Score = 72.8 bits (177), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/116 (31%), Positives = 63/116 (54%), Gaps = 5/116 (4%)
Query 9 SSSRKQCDFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYP 65
+ R DF +L AG + N +N V GP +T ++GG +++ + +DYP
Sbjct 5 ARKRLMRDFKRLQQDPPAGISGAPQDNNIMLWNAVIFGPDDTPWDGGTFKLSLQFSEDYP 64
Query 66 FSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNP 121
P++ F++++ HPN+ A GS+CLD++ W+P+Y + + + LL PNP
Sbjct 65 NKPPTVRFVSRMFHPNI-YADGSICLDILQNQWSPIYDVAAIL-TSIQSLLCDPNP 118
> At1g14400
Length=152
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/116 (31%), Positives = 63/116 (54%), Gaps = 5/116 (4%)
Query 9 SSSRKQCDFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYP 65
+ R DF +L AG + N +N V GP +T ++GG +++ + +DYP
Sbjct 5 ARKRLMRDFKRLQQDPPAGISGAPQDNNIMLWNAVIFGPDDTPWDGGTFKLSLQFSEDYP 64
Query 66 FSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNP 121
P++ F++++ HPN+ A GS+CLD++ W+P+Y + + + LL PNP
Sbjct 65 NKPPTVRFVSRMFHPNI-YADGSICLDILQNQWSPIYDVAAIL-TSIQSLLCDPNP 118
> At2g16740
Length=148
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP E+ Y GGV+ V++ P DYPF P + F K+ HPN++ ++G++CLD++
Sbjct 33 WQATIMGPNESPYSGGVFLVNIHFPPDYPFKPPKVVFRTKVFHPNIN-SNGNICLDILKD 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLTDPNPDD 117
> Hs4507773
Length=147
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y+GGV+ + V P DYPF P I F KI HPN++ ++GS+CLD++
Sbjct 33 WQATIMGPPDSAYQGGVFFLTVHFPTDYPFKPPKIAFTTKIYHPNIN-SNGSICLDILRS 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LL PNP D
Sbjct 92 QWSPALTVSKVL-LSICSLLCDPNPDD 117
> CE27822
Length=192
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 61/119 (51%), Gaps = 5/119 (4%)
Query 6 TQISSSRKQCDFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 62
T S R DF KL AG N + + GP+ET +E G +++ + +
Sbjct 2 TTPSRRRLMRDFKKLQEDPPAGVSGAPTEDNILTWEAIIFGPQETPFEDGTFKLSLEFTE 61
Query 63 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNP 121
+YP P++ F++K+ HPNV A GS+CLD++ W+P Y + + + LL PNP
Sbjct 62 EYPNKPPTVKFISKMFHPNV-YADGSICLDILQNRWSPTYDVAAIL-TSIQSLLDEPNP 118
> At5g56150
Length=148
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ + GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ +
Sbjct 33 WQATIMGPADSPFAGGVFLVTIHFPPDYPFKPPKVAFRTKVYHPNIN-SNGSICLDILKE 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 92 QWSPALTVSKVL-LSICSLLTDPNPDD 117
> Hs7657046
Length=225
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 55/93 (59%), Gaps = 2/93 (2%)
Query 30 NNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSV 89
N ++ D V GP+ T Y GG++R+ + L D+P S P FL KI HPNV A+G +
Sbjct 36 NEEDLTDLQVTIEGPEGTPYAGGLFRMKLLLGKDFPASPPKGYFLTKIFHPNVG-ANGEI 94
Query 90 CLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 122
C++V+ + WT + +V + + LL +PNP+
Sbjct 95 CVNVLKRDWTAELGIRHVL-LTIKCLLIHPNPE 126
> At3g08700
Length=149
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 50/87 (57%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y GGV+ V + DYPF P + F K+ HPN+D + GS+CLD++ +
Sbjct 34 WQATIMGPHDSPYSGGVFTVSIDFSSDYPFKPPKVNFKTKVYHPNID-SKGSICLDILKE 92
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P + V + + LLT PNP D
Sbjct 93 QWSPAPTTSKVL-LSICSLLTDPNPND 118
> CE03482
Length=147
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP E+ Y+GGV+ + + P DYPF P + F +I HPN++ ++GS+CLD++
Sbjct 33 WQATIMGPPESPYQGGVFFLTIHFPTDYPFKPPKVAFTTRIYHPNIN-SNGSICLDILRS 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LL PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLCDPNPDD 117
> SPBC1198.09
Length=160
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 47/82 (57%), Gaps = 0/82 (0%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ V GP ET YEGG W + + + + YP S PS+ F KI+HPN+ +G VC+D++
Sbjct 37 WKAVIEGPTETPYEGGQWVLDIHVHEGYPISPPSVYFQTKIVHPNISWTNGEVCMDILKT 96
Query 97 TWTPLYSLVNVFDVFLPQLLTY 118
W+P +SL + + L Y
Sbjct 97 HWSPAWSLQSACLAIISLLSNY 118
> Hs4885417
Length=200
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 59/106 (55%), Gaps = 6/106 (5%)
Query 8 ISSSRKQCDFAKLLMAG------YDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLP 61
I+ R + +F ++L + ++L ++N + GP +T YEGG +++ + +P
Sbjct 4 IAVQRIKREFKEVLKSEETSKNQIKVDLVDENFTELRGEIAGPPDTPYEGGRYQLEIKIP 63
Query 62 DDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNV 107
+ YPF+ P + F+ KI HPN+ +G++CLD++ W +L V
Sbjct 64 ETYPFNPPKVRFITKIWHPNISSVTGAICLDILKDQWAAAMTLRTV 109
> CE03946
Length=260
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 38/139 (27%), Positives = 65/139 (46%), Gaps = 17/139 (12%)
Query 1 STMAQTQISSSRKQ-CDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVT 59
S +A +++ K+ + + + AG +E+ N D GP+ T Y GG + + V
Sbjct 16 SNLALARVTRKCKEVANASDITEAGIHVEIKENNLMDIKGFIKGPEGTPYAGGTFEIKVD 75
Query 60 LPDDYPFSSPSI---------------GFLNKILHPNVDEASGSVCLDVINQTWTPLYSL 104
+P+ YPF P + F+ +I HPN+ +G++CLD++ WT +L
Sbjct 76 IPEHYPFEPPKVTEIIFHIRAFEYIQAKFVTRIWHPNISSQTGTICLDILKDKWTASLTL 135
Query 105 VNVFDVFLPQLLTYPNPQD 123
V + L +L P P D
Sbjct 136 RTVL-LSLQAMLCSPEPSD 153
> Hs8393719
Length=147
Score = 70.9 bits (172), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y+GGV+ + + P DYPF P + F KI HPN++ ++GS+CLD++
Sbjct 33 WQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTKIYHPNIN-SNGSICLDILRS 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LL PNP D
Sbjct 92 QWSPALTVSKVL-LSICSLLCDPNPDD 117
> SPBC119.02
Length=147
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y GGV+ + + P DYPF P + F +I HPN++ ++GS+CLD++
Sbjct 33 WQATIMGPADSPYAGGVFFLSIHFPTDYPFKPPKVNFTTRIYHPNIN-SNGSICLDILRD 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LLT PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLTDPNPDD 117
> 7294892
Length=199
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 58/108 (53%), Gaps = 1/108 (0%)
Query 1 STMAQTQISSSRKQC-DFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVT 59
+ MA ++I K+ +++ +EL N + + GP +T YEGG + + +
Sbjct 2 ANMAVSRIKREFKEVMRSEEIVQCSIKIELVNDSWTELRGEIAGPPDTPYEGGKFVLEIK 61
Query 60 LPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNV 107
+P+ YPF+ P + F+ +I HPN+ +G++CLD++ W +L V
Sbjct 62 VPETYPFNPPKVRFITRIWHPNISSVTGAICLDILKDNWAAAMTLRTV 109
> Hs4507777
Length=147
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y+GGV+ + + P DYPF P + F +I HPN++ ++GS+CLD++
Sbjct 33 WQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTRIYHPNIN-SNGSICLDILRS 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LL PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLCDPNPDD 117
> 7293378
Length=190
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 33/99 (33%), Positives = 54/99 (54%), Gaps = 2/99 (2%)
Query 24 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 83
G + +N + D + GP T Y G++RV +TL D+P + P FL KI HPNV
Sbjct 14 GIKVLINESDVTDIQALIDGPAGTPYAAGIFRVKLTLNKDFPLTPPKAYFLTKIFHPNV- 72
Query 84 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 122
A+G +C++ + + W P + ++ + + LL PNP+
Sbjct 73 AANGEICVNTLKKDWKPDLGIKHIL-LTIKCLLIVPNPE 110
> Hs4507775
Length=147
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y+GGV+ + + P DYPF P + F +I HPN++ ++GS+CLD++
Sbjct 33 WQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTRIYHPNIN-SNGSICLDILRS 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LL PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLCDPNPDD 117
> YBR082c
Length=148
Score = 70.5 bits (171), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 34/81 (41%), Positives = 50/81 (61%), Gaps = 2/81 (2%)
Query 43 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 102
GP ++ Y GGV+ + + P DYPF P I F KI HPN++ A+G++CLD++ W+P
Sbjct 40 GPADSPYAGGVFFLSIHFPTDYPFKPPKISFTTKIYHPNIN-ANGNICLDILKDQWSPAL 98
Query 103 SLVNVFDVFLPQLLTYPNPQD 123
+L V + + LLT NP D
Sbjct 99 TLSKVL-LSICSLLTDANPDD 118
> 7299919
Length=147
Score = 70.1 bits (170), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 37 FNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQ 96
+ GP ++ Y+GGV+ + + P DYPF P + F +I HPN++ ++GS+CLD++
Sbjct 33 WQATIMGPPDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTRIYHPNIN-SNGSICLDILRS 91
Query 97 TWTPLYSLVNVFDVFLPQLLTYPNPQD 123
W+P ++ V + + LL PNP D
Sbjct 92 QWSPALTISKVL-LSICSLLCDPNPDD 117
> YDR059c
Length=148
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 50/81 (61%), Gaps = 2/81 (2%)
Query 43 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 102
GP ++ Y GGV+ + + P DYPF P + F KI HPN++ +SG++CLD++ W+P
Sbjct 40 GPSDSPYAGGVFFLSIHFPTDYPFKPPKVNFTTKIYHPNIN-SSGNICLDILKDQWSPAL 98
Query 103 SLVNVFDVFLPQLLTYPNPQD 123
+L V + + LLT NP D
Sbjct 99 TLSKVL-LSICSLLTDANPDD 118
Lambda K H
0.319 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191192512
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40