bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3456_orf1
Length=265
Score E
Sequences producing significant alignments: (Bits) Value
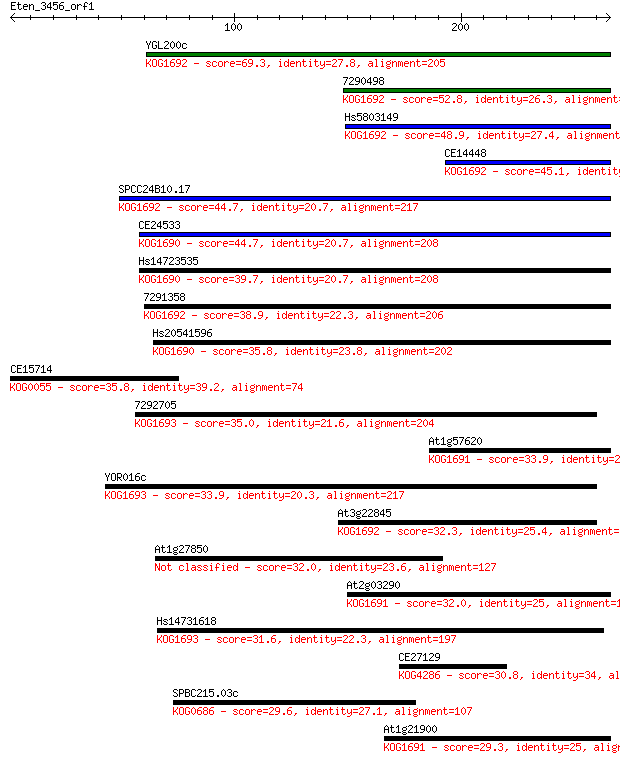
YGL200c 69.3 8e-12
7290498 52.8 8e-07
Hs5803149 48.9 1e-05
CE14448 45.1 2e-04
SPCC24B10.17 44.7 2e-04
CE24533 44.7 2e-04
Hs14723535 39.7 0.006
7291358 38.9 0.011
Hs20541596 35.8 0.092
CE15714 35.8 0.11
7292705 35.0 0.14
At1g57620 33.9 0.34
YOR016c 33.9 0.35
At3g22845 32.3 1.1
At1g27850 32.0 1.4
At2g03290 32.0 1.5
Hs14731618 31.6 1.7
CE27129 30.8 2.7
SPBC215.03c 29.6 7.6
At1g21900 29.3 7.8
> YGL200c
Length=203
Score = 69.3 bits (168), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 57/209 (27%), Positives = 91/209 (43%), Gaps = 23/209 (11%)
Query 61 LFFADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFGDGSIRVSVEGPVPSHRAA 120
LFF+ SA ++ LP R CFF S + E++ S+Q FGD P S +
Sbjct 14 LFFSASAHNVL--LPAYGRRCFFEDLS-KGDELSISFQ--FGD-------RNPQSSSQLT 61
Query 121 AAAAAQLPPLTPLFSSVEETAQLHAKL--PLRGEYAVCVRSLLSFEQT--VTIDFHLQTQ 176
P + +V +T+ L P +G + C + + +T VT + H
Sbjct 62 GDFIIYGPERHEVLKTVRDTSHGEITLSAPYKGHFQYCFLNENTGIETKDVTFNIHGVVY 121
Query 177 EKNSHPNQLALEDEALKLKGLMNEVLQKANSLFEQQSHAMVRLGVHAELGESTRRRATLW 236
PN L+ KL L EV ++QS+ ++R H EST R W
Sbjct 122 VDLDDPNTNTLDSAVRKLSKLTREVK-------DEQSYIVIRERTHRNTAESTNDRVKWW 174
Query 237 KSIQMGMQLVLTALQIYFVKSYFEVKTIV 265
Q+G+ + + QIY+++ +FEV ++V
Sbjct 175 SIFQLGVVIANSLFQIYYLRRFFEVTSLV 203
> 7290498
Length=208
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 58/120 (48%), Gaps = 2/120 (1%)
Query 148 PLRGEYAVCVRSLLSFEQTVTIDFHLQTQEKNSHPNQLALEDEA--LKLKGLMNEVLQKA 205
P +G Y VC + S + F + + E+E KL+ ++ E+
Sbjct 89 PAKGTYTVCFNNERSSMTPKLVMFSIDVGDAPQRAPGAPGEEEVGHTKLEDMIRELSGTL 148
Query 206 NSLFEQQSHAMVRLGVHAELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYFEVKTIV 265
S+ +Q + VR +H + E+T R LW + + + +++T Q+Y++K +FEVK +V
Sbjct 149 TSVKHEQEYMHVRDKIHRSVNENTNSRVVLWSTFEALVLVLMTVGQVYYLKRFFEVKRVV 208
> Hs5803149
Length=201
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 60/119 (50%), Gaps = 5/119 (4%)
Query 149 LRGEYAVCVRSLLSFEQTVTIDFHLQTQEKNSHPNQLALEDEA--LKLKGLMNEVLQKAN 206
+ G Y C + +S T+T + T + P +E EA KL+ ++NE+
Sbjct 86 MDGTYKFCFSNRMS---TMTPKIVMFTIDIGEAPKGQDMETEAHQNKLEEMINELAVAMT 142
Query 207 SLFEQQSHAMVRLGVHAELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYFEVKTIV 265
++ +Q + VR +H + ++T R LW + + + +T QIY++K +FEV+ +V
Sbjct 143 AVKHEQEYMEVRERIHRAINDNTNSRVVLWSFFEALVLVAMTLGQIYYLKRFFEVRRVV 201
> CE14448
Length=203
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 43/73 (58%), Gaps = 0/73 (0%)
Query 193 KLKGLMNEVLQKANSLFEQQSHAMVRLGVHAELGESTRRRATLWKSIQMGMQLVLTALQI 252
KL+ ++ E+ S+ +Q + VR VH + E+T R +W + + + + +T QI
Sbjct 131 KLEEMVRELSSALMSVKHEQEYMEVRERVHRNINENTNSRVVMWAAFEAFVLVGMTVGQI 190
Query 253 YFVKSYFEVKTIV 265
+++K +FEV+T+V
Sbjct 191 FYLKRFFEVRTMV 203
> SPCC24B10.17
Length=199
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 45/222 (20%), Positives = 90/222 (40%), Gaps = 28/222 (12%)
Query 49 LLLFSFFFSLKILFFADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFG-DGSIR 107
+ F+ F ++ +F L P RECF+ + + +++ +YQ+ G D +
Sbjct 1 MAFFNVFKAVLCAYFISVVFGHGITLKPHQRECFYENLRNND-QMSVTYQTNVGGDQLVS 59
Query 108 VSVEGPVPSHRAAAAAAAQLPPLTPLFSSVEETAQLHAKLPLRGEYAVCVRSLLSFEQTV 167
+S+ P A ++P AQ + G+Y C + ++
Sbjct 60 MSIYNP-----AGQIMHQEVP---------NSMAQYSFTVKNPGKYMYCFYNDALDGESK 105
Query 168 TIDFHLQTQEKNSHPNQLALEDEALK----LKGLMNEVLQKANSLFEQQSHAMVRLGVHA 223
+ F++ + + DE L L G + ++ + + +Q + + R +H
Sbjct 106 EVLFNVH--------GVIYISDEDLDANNPLLGKVRQLHDTISKVKHEQEYFVARERIHR 157
Query 224 ELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYFEVKTIV 265
EST R W +Q + + + QI+++K FEVK +V
Sbjct 158 NTAESTNDRVKWWSILQTVILVSVCVFQIFYLKRLFEVKRVV 199
> CE24533
Length=211
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 43/211 (20%), Positives = 95/211 (45%), Gaps = 7/211 (3%)
Query 58 LKILFFADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFGDGSIRVSVEGPVPSH 117
+ +L A A ++ F + ++CF + D E+ +TG+Y+ D + + + P
Sbjct 5 IALLSLATYADSLYFHIAETEKKCFIEEIPD-ETMVTGNYKVQLYDPNTKGYGDYPNIGM 63
Query 118 RAAAAAAAQLPPLTPLFSSVEETAQLHAKLPLRGEYAVCVRSL-LSFEQTVTIDFHLQTQ 176
L+ L+++ E + P GE+ +C+ S ++ + HL Q
Sbjct 64 HVEVKDPEDKVILSKLYTA-EGRFTFTSNTP--GEHVICIYSNSTAWFNGAQLRVHLDIQ 120
Query 177 EKNSHPN--QLALEDEALKLKGLMNEVLQKANSLFEQQSHAMVRLGVHAELGESTRRRAT 234
+ + Q+A +D+ +L+ + ++L + + + ++Q++ R + EST R
Sbjct 121 AGDHAQDYAQIAQKDKLNELQLRIRQLLDQVDQITKEQNYQRYREERFRQTSESTNSRVF 180
Query 235 LWKSIQMGMQLVLTALQIYFVKSYFEVKTIV 265
W Q+ + + A Q+ ++ +FE K +V
Sbjct 181 YWSIAQVVVLAITGAWQMRHLRGFFEAKKLV 211
> Hs14723535
Length=214
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 43/229 (18%), Positives = 92/229 (40%), Gaps = 40/229 (17%)
Query 58 LKILFFADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFGDGSIRVSVEGPVPSH 117
L +L+ A +A+ F + ++CF + D E+ + G+Y++ D
Sbjct 5 LLVLWLATRGSALYFHIGETEKKCFIEEIPD-ETMVIGNYRTQLYD-------------- 49
Query 118 RAAAAAAAQLPPLTPLFSSVEETAQLHAKLPLR----------------GEYAVCVRSL- 160
+ P TP E K+ L GE+ +C+ S
Sbjct 50 ----KQREEYQPATPGLGMFVEVKDPEDKVILARQYGSEGRFTFTSHTPGEHQICLHSNS 105
Query 161 ----LSFEQTVTIDFHLQTQEKNSHPNQLALEDEALKLKGLMNEVLQKANSLFEQQSHAM 216
L + + +Q E + ++A +D+ +L+ + +++++ + ++Q++
Sbjct 106 TKFSLFAGGMLRVHLDIQVGEHANDYAEIAAKDKLSELQLRVRQLVEQVEQIQKEQNYQR 165
Query 217 VRLGVHAELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYFEVKTIV 265
R + EST +R W +Q + + + Q+ +KS+FE K +V
Sbjct 166 WREERFRQTSESTNQRVLWWSILQTLILVAIGVWQMRHLKSFFEAKKLV 214
> 7291358
Length=203
Score = 38.9 bits (89), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 46/220 (20%), Positives = 95/220 (43%), Gaps = 38/220 (17%)
Query 60 ILFFADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFGD-GSIRVSVEGPVPSHR 118
+L +A + L CF+ ++ ++T S++ G + V + GP R
Sbjct 8 VLVLFQAACGFIVTLDAHETMCFY-DHANVSDKVTVSFEVMEGGFKDVGVEIAGP-DDDR 65
Query 119 AAAAAAAQLPPLTPLFSSVEETAQLHAKLPLRGEYAVCVRSLLS----------FEQTVT 168
+ + T F++++E G Y +C + +S F
Sbjct 66 LHHSKQDTMGSFT--FTAMKE-----------GRYQLCFDNKMSTMTPKILMFQFHVARA 112
Query 169 IDFHLQTQEKNSHPNQLALED--EALKLKGLMNEVLQKANSLFEQQSHAMVRLGVHAELG 226
I+F++ + ++ ++D E ++ ++N++ K ++ +Q + R H E+
Sbjct 113 IEFYMDSSKR--------VDDVIEQATVQSMINQLSAKLGAVKMEQEYMHFRYRGHLEVS 164
Query 227 ESTRRRATLWKSIQMGMQLVLTA-LQIYFVKSYFEVKTIV 265
+ R W SI M L++TA L++Y++K +FEVK +V
Sbjct 165 DMVELRVLAW-SIFGPMMLIITAVLEVYYLKHFFEVKRVV 203
> Hs20541596
Length=227
Score = 35.8 bits (81), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 48/213 (22%), Positives = 93/213 (43%), Gaps = 20/213 (9%)
Query 64 ADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFGDGSIRVSVEGPVPSHRAAAAA 123
A A + F + + CF + D E+ + G+Y++ D V + PS
Sbjct 24 ATGAQGLYFHIGETEKRCFIEEIPD-ETMVIGNYRTQMWDKQKEVFL----PSTPGLGMH 78
Query 124 AAQLPPLTPLFSSVEETAQ----LHAKLPLRGEYAVCVRS---LLSFEQTVTIDFHLQTQ 176
P + S + ++ + P G++ +C+ S ++ + HL Q
Sbjct 79 VEVKDPDGKVVLSRQYGSEGRFTFTSHTP--GDHQICLHSNSTRMALFAGGKLRVHLDIQ 136
Query 177 ---EKNSHPNQLALEDEALKLKGLMNEVLQKANSLFEQQSHAMVRLGVHAELGESTRRRA 233
N++P ++A +D+ +L+ ++L + + ++Q + R EST +R
Sbjct 137 VGEHANNYP-EIAAKDKLTELQLRARQLLDQVEQIQKEQDYQRYREERFRLTSESTNQRV 195
Query 234 TLWKSIQMGMQLVLTAL-QIYFVKSYFEVKTIV 265
LW SI + L+LT + Q+ +KS+FE K +V
Sbjct 196 -LWWSIAQTVILILTGIWQMRHLKSFFEAKKLV 227
> CE15714
Length=1294
Score = 35.8 bits (81), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 43/85 (50%), Gaps = 18/85 (21%)
Query 1 ILRARFLEFAHSVFSCSFFGYFASF-FSLFLIFYSGITMSPRSRCFSLLLLLFSFFFSLK 59
I+R FA+S+ FF Y A+F F LFLIF + M P ++L +LF+ FS
Sbjct 930 IIRGLTYGFANSI---QFFTYAAAFRFGLFLIFDKNVLMEPE----NVLRVLFAISFSFG 982
Query 60 ILFFADS----------AAAIMFDL 74
+ FA S AA ++F++
Sbjct 983 TIGFAASYFPEYIKATFAAGLIFNM 1007
> 7292705
Length=210
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 44/210 (20%), Positives = 88/210 (41%), Gaps = 26/210 (12%)
Query 56 FSLKILFFADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFGDGSIRVSVEGPVP 115
F++ ++ SA FDL A +CF+ EI + ++F +VS G +
Sbjct 8 FAVALMMHCISAVEFTFDLADNAVDCFY-------EEIKKNSSAYF---EFQVSAGGQLD 57
Query 116 SHRAAAAAAAQLPPLTPLFSSVEETAQLHAKLP-LRGEYAVCVRSLLSF--EQTVTIDFH 172
+ P ++S + T H + G Y C + S + V +DF
Sbjct 58 -----VDVTLKDPQGKVIYSLEKATFDSHQFVAETTGVYTACFGNQFSAFSHKIVYVDF- 111
Query 173 LQTQEKNSHPNQLALEDEALKLKGL--MNEVLQKA-NSLFEQQSHAMVRLGVHAELGEST 229
Q E+ + P +++ A L + ++ + K N + + Q+H +R + E
Sbjct 112 -QVGEEPALP---GVDEHATVLTQMETSSQAIHKGLNDILDAQTHHRLREAQGRKRAEDL 167
Query 230 RRRATLWKSIQMGMQLVLTALQIYFVKSYF 259
+R +W S++ +V+ +QI ++++F
Sbjct 168 NQRVMVWSSLETAAVIVIGLVQIMVLRNFF 197
> At1g57620
Length=212
Score = 33.9 bits (76), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 43/80 (53%), Gaps = 7/80 (8%)
Query 186 ALEDEALKLKGLMNEVLQKANSLFEQQSHAMVRLGVHAELGESTRRRATLWKSIQMGMQL 245
+E E KL+G + + + N ++ + A +R+ + E T R + + +G+ +
Sbjct 140 GVELEFKKLEGAVEAIHE--NLIYLRNREAEMRI-----VSEKTNSRVAWYSIMSLGICI 192
Query 246 VLTALQIYFVKSYFEVKTIV 265
V++ LQI ++K YFE K ++
Sbjct 193 VVSGLQILYLKQYFEKKKLI 212
> YOR016c
Length=207
Score = 33.9 bits (76), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 44/222 (19%), Positives = 87/222 (39%), Gaps = 29/222 (13%)
Query 43 RCFSLLLLLFSFFFSLKILFFADSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFG 102
R F+L+ +LFS SL F+ + A + LP +EC + S + + SYQ G
Sbjct 2 RVFTLIAILFSS--SLLTHAFSSNYAPVGISLPAFTKECLYYDLSSDKDVLVVSYQVLTG 59
Query 103 DGSIRVSVEGPVPSHRAAAAAAAQLPPLTPLFSSVEETAQLHAKLPLR----GEYAVCV- 157
G+ + + P V E + H+ L+ G+Y C+
Sbjct 60 -GNFEIDFDITAPDGSVI----------------VTERQKKHSDFLLKSFGIGKYTFCLS 102
Query 158 RSLLSFEQTVTIDFHLQTQEKNSHPNQLALEDEALKLKGLMNEVLQKANSLFEQQSHAMV 217
+ + + V I + + +SH ++ E + + E+ + N + + +
Sbjct 103 NNYGTSPKKVEITLEKEKEIVSSHESK-----EDIIANNAIEEIDRNLNKITKTMDYLRA 157
Query 218 RLGVHAELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYF 259
R + ST R T + MG+ + ++ +Q ++ +F
Sbjct 158 REWRNMYTVSSTESRLTWLSLLIMGVMVGISIVQALIIQFFF 199
> At3g22845
Length=214
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 50/117 (42%), Gaps = 9/117 (7%)
Query 146 KLPLRGEYAVCVRSLLSFEQTVTIDFHLQTQEKNSH-PNQLAL-EDEALK-LKGLMNEVL 202
K P G Y C + S +TV+ H+ H PN+ L +DE L + + E+
Sbjct 95 KAPKSGMYKFCFHNPYSTPETVSFYIHV------GHIPNEHDLAKDEHLDPVNVKIAELR 148
Query 203 QKANSLFEQQSHAMVRLGVHAELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYF 259
+ S+ +Q + R H ESTR+R + + + LQ+ +++ F
Sbjct 149 EALESVVAEQKYLKARDTRHRHTNESTRKRVIFYTVGEYIFLAAASGLQVLYIRKLF 205
> At1g27850
Length=1148
Score = 32.0 bits (71), Expect = 1.4, Method: Composition-based stats.
Identities = 30/134 (22%), Positives = 60/134 (44%), Gaps = 16/134 (11%)
Query 65 DSAAAIMFDLPPQARECFFVKCSDQESEITGSYQSFFGDGSIRVSVEGPVPSHRAAAAAA 124
D A+ ++ + R+ F ++ SD ++ + F + +I V E S R A
Sbjct 36 DDDLALFSEMQDKERDSFLLQSSDDLEDVFSTKLKHFSEFTIPVQGE----SSRLLTAEG 91
Query 125 AQ-------LPPLTPLFSSVEETAQLHAKLPLRGEYAVCVRSLLSFEQTVTIDFHLQTQE 177
+ PP TPLF S+++ A + RG +S +S ++ T++ ++ +
Sbjct 92 DKNDYDWLLTPPDTPLFPSLDDQPPA-ASVVRRGR----PQSQISLSRSSTMEKSRRSSK 146
Query 178 KNSHPNQLALEDEA 191
++ PN+L+ A
Sbjct 147 GSASPNRLSTSPRA 160
> At2g03290
Length=213
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 29/121 (23%), Positives = 55/121 (45%), Gaps = 9/121 (7%)
Query 150 RGEYAVCVRSL-LSFEQTVTIDFHLQTQEKNSHPNQLALEDEA----LKLKGLMNEVLQK 204
G Y C+ ++ E T+TIDF +T + +A + + ++K LM+ V+
Sbjct 97 NGSYVACITAIDYKPETTLTIDFDWKTGVHSKEWTNVAKKSQVDMMEYQVKTLMDTVI-- 154
Query 205 ANSLFEQQSHAMVRLGVHAELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYFEVKTI 264
S+ E+ + R EL ST + + + L + LQ + +K++FE K +
Sbjct 155 --SIHEEMYYLREREEEMQELNRSTNSKMAWLSFGSLVVCLSVAGLQFWHLKTFFEKKKL 212
Query 265 V 265
+
Sbjct 213 I 213
> Hs14731618
Length=224
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 44/206 (21%), Positives = 75/206 (36%), Gaps = 32/206 (15%)
Query 66 SAAAIMFDLPPQARECFF------VKCSDQESEITGSYQSFFGDGSIRVSVEGPVPSHRA 119
A+ I F+LP A++CF+ KC+ + ITG G + +E P
Sbjct 33 GASEITFELPDNAKQCFYEDIAQGTKCTLEFQVITG------GHYDVDCRLEDP------ 80
Query 120 AAAAAAQLPPLTPLFSSVEETAQLHAKLPLRGEYAVCVRSLLSFEQTVTIDFHLQTQEKN 179
+ + S TA + G Y C + S T+ F Q E
Sbjct 81 --DGKVLYKEMKKQYDSFTFTASKN------GTYKFCFSNEFSTFTHKTVYFDFQVGED- 131
Query 180 SHPNQLALEDEALKLKGLMN---EVLQKANSLFEQQSHAMVRLGVHAELGESTRRRATLW 236
P E+ L + + + + S+ + Q+H +R E R W
Sbjct 132 --PPLFPSENRVSALTQMESACVSIHEALKSVIDYQTHFRLREAQGRSRAEDLNTRVAYW 189
Query 237 KSIQMGMQLVLTALQIYFVKSYFEVK 262
+ + LV++ Q++ +KS+F K
Sbjct 190 SVGEALILLVVSIGQVFLLKSFFSDK 215
> CE27129
Length=3674
Score = 30.8 bits (68), Expect = 2.7, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 173 LQTQEKNSHPNQLALEDEALKLKGLMNEVLQKANSLFEQQSHAMVRL 219
+Q +E+ + N+L LE E L + L L++AN FEQ + R+
Sbjct 741 IQIKEQEARVNRLQLELEHLHVAKLNARQLKRANDAFEQFAKGWARI 787
> SPBC215.03c
Length=422
Score = 29.6 bits (65), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 43/109 (39%), Gaps = 28/109 (25%)
Query 73 DLPPQARECFFVKCSDQESEITGSYQSFFGDGSIRVSVEGPVPSHRAAAAAAAQLPPLTP 132
DL P R C KC+ + S + + Q D S+ + + P LT
Sbjct 259 DLDPSLRRCLQAKCNRKYSLLLDTLQQNAQDYSLDMYL----------------APQLTN 302
Query 133 LFSSVEETAQLHAKLPLRGEYAVCVRSLLSFEQTVTIDFHLQTQ--EKN 179
LFS + E + L +P S L F + + +DFH+ EKN
Sbjct 303 LFSLIRERSLLDYLIPY---------SALPFSK-IAVDFHIDENFIEKN 341
> At1g21900
Length=216
Score = 29.3 bits (64), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 49/102 (48%), Gaps = 6/102 (5%)
Query 166 TVTIDFHLQTQEKN--SHPNQLALEDEALKLKGLMNEVLQKANSLFEQQSHAMVRLGVHA 223
T+ +D+ + K+ S + +E L+L+ L VL S+ E ++ R
Sbjct 119 TLGVDWKMGIAAKDWDSVAKKEKIEGVELQLRRLEGLVL----SIRENLNYIKDREAEMR 174
Query 224 ELGESTRRRATLWKSIQMGMQLVLTALQIYFVKSYFEVKTIV 265
E+ E+T R + + +G+ +V+ QI ++K YF K ++
Sbjct 175 EVSETTNSRVAWFSIMSLGVCVVVVGSQILYLKRYFHKKKLI 216
Lambda K H
0.327 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5649423594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40