bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3445_orf1
Length=138
Score E
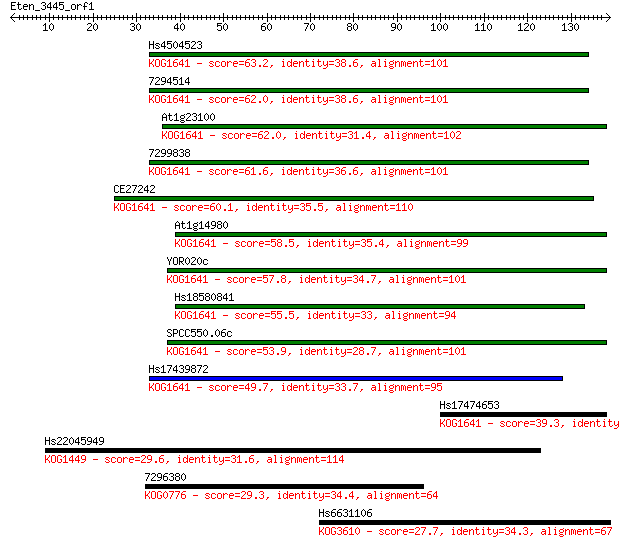
Sequences producing significant alignments: (Bits) Value
Hs4504523 63.2 2e-10
7294514 62.0 4e-10
At1g23100 62.0 4e-10
7299838 61.6 5e-10
CE27242 60.1 1e-09
At1g14980 58.5 4e-09
YOR020c 57.8 6e-09
Hs18580841 55.5 3e-08
SPCC550.06c 53.9 9e-08
Hs17439872 49.7 2e-06
Hs17474653 39.3 0.002
Hs22045949 29.6 2.0
7296380 29.3 2.1
Hs6631106 27.7 7.0
> Hs4504523
Length=102
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 57/102 (55%), Gaps = 5/102 (4%)
Query 33 MSGLAAR-LMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSPPKGTGNS 91
M+G A R +PLF R +V++ TV+KGGI +P + K A VVAV
Sbjct 1 MAGQAFRKFLPLFDRVLVERSAAETVTKGGIMLPEKSQGK--VLQATVVAVGSGSKGKGG 58
Query 92 AVSEDWAKLQVGQTVMVPEFGGMKVEVDDEELFVFRGEDILA 133
+ ++VG V++PE+GG KV +DD++ F+FR DIL
Sbjct 59 EIQP--VSVKVGDKVLLPEYGGTKVVLDDKDYFLFRDGDILG 98
> 7294514
Length=103
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 59/104 (56%), Gaps = 7/104 (6%)
Query 33 MSGLAARLMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSPPKGTGNSA 92
M+ +++P+ R ++Q+ + T +KGGI +P A K V+AV P GT N++
Sbjct 1 MAAAIKKIIPMLDRILIQRAEALTKTKGGIVLPEKAVGK--VLEGTVLAVGP--GTRNAS 56
Query 93 VSEDW-AKLQVGQTVMVPEFGGMKV--EVDDEELFVFRGEDILA 133
++ G V++PEFGG KV E D +ELF+FR DILA
Sbjct 57 TGNHIPIGVKEGDRVLLPEFGGTKVNLEGDQKELFLFRESDILA 100
> At1g23100
Length=97
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 60/102 (58%), Gaps = 5/102 (4%)
Query 36 LAARLMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSPPKGTGNSAVSE 95
+A RL+P R +V+K+ P + + GI +P +S + +V+AV P G + A +
Sbjct 1 MAKRLIPTLNRVLVEKILPPSKTVSGILLPEKSS---QLNSGRVIAVGP--GARDRAGNL 55
Query 96 DWAKLQVGQTVMVPEFGGMKVEVDDEELFVFRGEDILAVVKD 137
++ G V++PEFGG +V++ ++E ++R EDI+A + +
Sbjct 56 IPVSVKEGDNVLLPEFGGTQVKLGEKEFLLYRDEDIMATLHE 97
> 7299838
Length=102
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 37/106 (34%), Positives = 61/106 (57%), Gaps = 12/106 (11%)
Query 33 MSGLAARLMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSP----PKGT 88
MS + +++P+ R ++Q+ + T + GGI +P + PK VVAV P P G
Sbjct 1 MSNVIKKVIPMLDRILIQRFEVKTTTAGGILLPEESVPKE--MQGVVVAVGPGARNPAGA 58
Query 89 GNSAVSEDWAKLQVGQTVMVPEFGGMKVEVDDEELFV-FRGEDILA 133
G+ +V ++ G V++P++GG KV++DD+ +V FR DILA
Sbjct 59 GHLSVG-----VKEGDRVLLPKYGGTKVDMDDKREYVLFRESDILA 99
> CE27242
Length=108
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 58/110 (52%), Gaps = 5/110 (4%)
Query 25 FLTAVPAKMSGLAARLMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSP 84
FLTAV + S + PL+ R +V++V T +KGGI +P KS + + VS
Sbjct 2 FLTAV-RRSSNVLKTFKPLYDRVLVERVAAETKTKGGIMLPE----KSQGKVLEATVVSA 56
Query 85 PKGTGNSAVSEDWAKLQVGQTVMVPEFGGMKVEVDDEELFVFRGEDILAV 134
G N ++ G V++PE+GG KV V+D+E +FR D+L V
Sbjct 57 GAGLRNEKGELVALTVKPGDRVLLPEYGGTKVVVEDKEYSIFRESDLLGV 106
> At1g14980
Length=97
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 59/100 (59%), Gaps = 7/100 (7%)
Query 39 RLMPLFGRCVVQKV-QPATVSKGGIYIPASASPKSGCHMAKVVAVSPPKGTGNSAVSEDW 97
RL+P F R +VQ+V QPA ++ GI +P +S + KV+AV P G+ +
Sbjct 3 RLIPTFNRILVQRVIQPAK-TESGILLPEKSSK---LNSGKVIAVGP--GSRDKDGKLIP 56
Query 98 AKLQVGQTVMVPEFGGMKVEVDDEELFVFRGEDILAVVKD 137
++ G TV++PE+GG +V++ + E +FR ED+L + +
Sbjct 57 VSVKEGDTVLLPEYGGTQVKLGENEYHLFRDEDVLGTLHE 96
> YOR020c
Length=106
Score = 57.8 bits (138), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 61/105 (58%), Gaps = 10/105 (9%)
Query 37 AARLMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSP--PKGTGNSAVS 94
A ++PL R +VQ+++ + G+Y+P K + A+VVAV P GN V
Sbjct 8 AKSIVPLMDRVLVQRIKAQAKTASGLYLPEKNVEK--LNQAEVVAVGPGFTDANGNKVVP 65
Query 95 EDWAKLQVGQTVMVPEFGGMKVEV-DDEELFVFRGEDILA-VVKD 137
+ ++VG V++P+FGG +++ +D+E+ +FR +ILA + KD
Sbjct 66 Q----VKVGDQVLIPQFGGSTIKLGNDDEVILFRDAEILAKIAKD 106
> Hs18580841
Length=91
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 50/94 (53%), Gaps = 15/94 (15%)
Query 39 RLMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSPPKGTGNSAVSEDWA 98
+ +PL R +V++ TV+KGGI +P + K A+ + P
Sbjct 8 KFLPLLDRVLVERRAAETVTKGGIMLPEKSQGK--LLQARGREIQP-------------V 52
Query 99 KLQVGQTVMVPEFGGMKVEVDDEELFVFRGEDIL 132
++VG V++PE+GG KV +DD++ F+FR DIL
Sbjct 53 SVKVGDKVLLPEYGGTKVVLDDKDYFLFRDGDIL 86
> SPCC550.06c
Length=104
Score = 53.9 bits (128), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 56/101 (55%), Gaps = 4/101 (3%)
Query 37 AARLMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSPPKGTGNSAVSED 96
A ++PL R +VQ+++ T + GI++P + K +V++V KG N
Sbjct 8 AKSIVPLLDRILVQRIKADTKTASGIFLPEKSVEK--LSEGRVISVG--KGGYNKEGKLA 63
Query 97 WAKLQVGQTVMVPEFGGMKVEVDDEELFVFRGEDILAVVKD 137
+ VG V++P +GG ++V +EE ++R ++LA++K+
Sbjct 64 QPSVAVGDRVLLPAYGGSNIKVGEEEYSLYRDHELLAIIKE 104
> Hs17439872
Length=132
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 51/96 (53%), Gaps = 5/96 (5%)
Query 33 MSGLAAR-LMPLFGRCVVQKVQPATVSKGGIYIPASASPKSGCHMAKVVAVSPPKGTGNS 91
M+G A R + LF R V+++ TV+KGGI +P + K AK VAV
Sbjct 1 MAGQAFRKFLLLFDRVFVERIATTTVTKGGIMLPEKSQGK--VLQAKGVAVGSGSKGKGG 58
Query 92 AVSEDWAKLQVGQTVMVPEFGGMKVEVDDEELFVFR 127
+ ++VG ++ E+GG +V +DD++ F+FR
Sbjct 59 EIQP--VSMRVGDKFLLLEYGGTRVVLDDKDYFLFR 92
> Hs17474653
Length=174
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 100 LQVGQTVMVPEFGGMKVEVDDEELFVFRGEDILAVVKD 137
++VG V++PE+ G+KV +DD++ F FR +IL D
Sbjct 137 VKVGDKVLLPEYRGIKVSLDDKDSFFFRDNNILGKYLD 174
> Hs22045949
Length=1444
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 51/124 (41%), Gaps = 13/124 (10%)
Query 9 SDNASWYLSSSYLFLRFLTAVPAK-MSGLAARLMPLFGRCVVQKVQPATVSKGGIYIPAS 67
S N S ++ L + T PAK M L + +P+ G+ TV+ GG +IPA+
Sbjct 322 SRNGSVFVRGQRLSVEKATIRPAKSMDSLCS--VPVEGKETKGNFN-RTVTTGGFFIPAT 378
Query 68 ASPKSG----CHMAKVVAV-----SPPKGTGNSAVSEDWAKLQVGQTVMVPEFGGMKVEV 118
+G C + K PP G VS D + LQ Q PE + V
Sbjct 379 KMHSTGTGSSCDLTKQEGEWGQEGMPPGAEGGFDVSSDRSHLQGAQARPPPEQLKVFRPV 438
Query 119 DDEE 122
+D E
Sbjct 439 EDPE 442
> 7296380
Length=448
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 32/71 (45%), Gaps = 9/71 (12%)
Query 32 KMSGLAARLMPLFGRCVVQKVQPATVSKGGIYIPA-------SASPKSGCHMAKVVAVSP 84
+ SGL R+M R + ++QP V+K + S + SG H + V P
Sbjct 5 RASGL--RIMQQMRRRIPVELQPLQVAKAAPALQTFTSQRWTSTTTTSGKHASPQVTTPP 62
Query 85 PKGTGNSAVSE 95
P+ N AVSE
Sbjct 63 PRHDWNRAVSE 73
> Hs6631106
Length=2135
Score = 27.7 bits (60), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 5/71 (7%)
Query 72 SGCHMAKVVAVSPPKGTGNSAVSEDWA----KLQVGQTVMVPEFGGMKVEVDDEELFVFR 127
SG +A AV P G G D+A K+ P GG ++ ++ +L R
Sbjct 1133 SGEEVAGATAVEVP-GRGRGVSEHDFAYQDPKVHSIFPARGPRAGGTRLTLNGSKLLTGR 1191
Query 128 GEDILAVVKDQ 138
EDI VV DQ
Sbjct 1192 LEDIRVVVGDQ 1202
Lambda K H
0.317 0.132 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1498437086
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40